Eulor10
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000122 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 317 |
| Kimura value | 25.37 |
| Tau index | 0.0000 |
| Description | Eulor10 (Euteleostomi-conserved low frequency repeat 10) |
| Comment | Putative DNA transposon (assignment unclear). Termini unclear. Eulor10 is present in mammals and birds (~40 copies), and alligator (~100 copies). Follows characteristic hairpin-tail structure, with at least two internal palindromes. |
| Sequence |
TGAAACGCGGGNCGGATTCATCAAAGGAGAGCTGGNANTTACCGACTCCGNATATTTTGCTTNAATTTTTTTTNTCTGATTCATGAANCGCTTTTCGAAATTCGAAAAGCGGTTCATGAATCGNTCGGGAGCCGGCAAAAATTAATAGTAATGAGCTCATTTCCATAGAAATGGGCTCATTACCATGCCGGCTGCCGAAAATTTACGAGGCCGGNTTCTTGCCGGCAAACGGGGTCCTGCAGGCAGTGTCCTTCCGCCTGCCTGCAGGAAATAATCCCNTNTTNAAAAATTAANCAAAAACAAATTTTAGTTTATTT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor10 | TCF7L1 | 16 | 27 | + | 9.62 | ATTCATCAAAGG |
| Eulor10 | Zm00001d049364 | 187 | 197 | - | 9.39 | CGGCAGCCGGC |
| Eulor10 | opa | 256 | 267 | + | 9.05 | GCCTGCCTGCAG |
| Eulor10 | TCX6 | 93 | 107 | + | 8.84 | TTTCGAAATTCGAAA |
| Eulor10 | ceh-38 | 137 | 150 | + | 8.04 | AAAAATTAATAGTA |
| Eulor10 | ZNF460 | 250 | 265 | + | 5.40 | CCTTCCGCCTGCCTGC |
| Eulor10 | RFX2 | 159 | 172 | - | 5.17 | ATTTCTATGGAAAT |
| Eulor10 | RFX2 | 159 | 172 | + | 5.10 | ATTTCCATAGAAAT |
| Eulor10 | IXR1 | 100 | 114 | + | 0.95 | ATTCGAAAAGCGGTT |
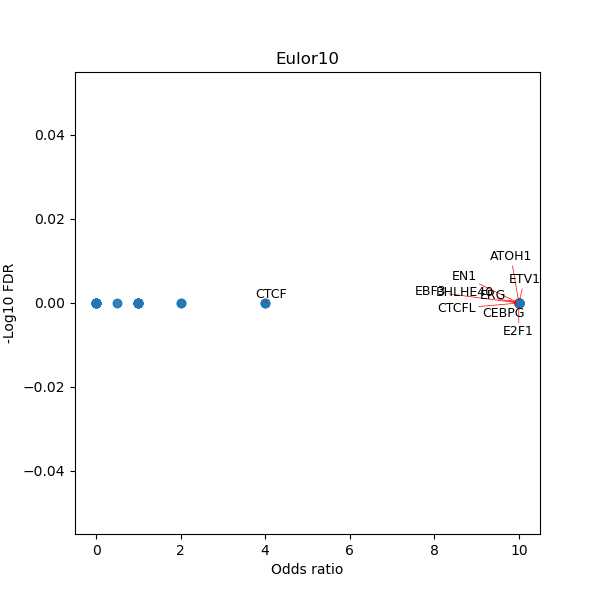
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.