Eulor1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000121 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 357 |
| Kimura value | 24.84 |
| Tau index | 0.0000 |
| Description | Eulor1 (Euteleostomi-conserved low frequency repeat 1) |
| Comment | Eulor1 forms a near-perfect hairpin and is present in the chicken (~80 copies) and mammalian genomes (100-150 copies). It may represent an incomplete non-autonomous DNA transposon or a mixture of related DNA transposons of different length. |
| Sequence |
CAGCAGGCCGGATTCATCAAAAGGATAACGGGTAGATATTTTCCGTTTGTNGAATTTTAACGAATAAACGGCATTTCTATTCGTTATTTATCTACTTTCGAATTTTAACGAATAGTTCTAGTGATAATTACCGAATTTCTATATTTATAGAAAACCGGCACTTCATAAATATCGAATTGTGCTATTATCTACATATGTGCCGGTTTTCTATAAATATAGAAATTCGGTAATTATCACTAGAACTATTCGTTAAAATTCGAAAGTAGATAAATAACGAATAGGAATGCCATTTATTCGTTAAAATTCTACAAAANAAAAATATCTNCCCGTTATCCCTTTGATGAATTCGGCCCATTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor1 | PK02532.1 | 317 | 324 | + | 14.19 | AAATATCT |
| Eulor1 | PK02532.1 | 34 | 41 | - | 14.19 | AAATATCT |
| Eulor1 | Lef1 | 335 | 342 | + | 14.17 | CCTTTGAT |
| Eulor1 | AP1 | 136 | 148 | - | 14.06 | ATAAATATAGAAA |
| Eulor1 | AP1 | 210 | 222 | + | 14.06 | ATAAATATAGAAA |
| Eulor1 | Crg-1 | 264 | 274 | + | 13.97 | TAGATAAATAA |
| Eulor1 | Crg-1 | 84 | 94 | - | 13.97 | TAGATAAATAA |
| Eulor1 | Zic1::Zic2 | 1 | 7 | + | 13.93 | CAGCAGG |
| Eulor1 | TCX2 | 248 | 262 | + | 13.90 | CGTTAAAATTCGAAA |
| Eulor1 | TCX2 | 96 | 110 | - | 13.90 | CGTTAAAATTCGAAA |
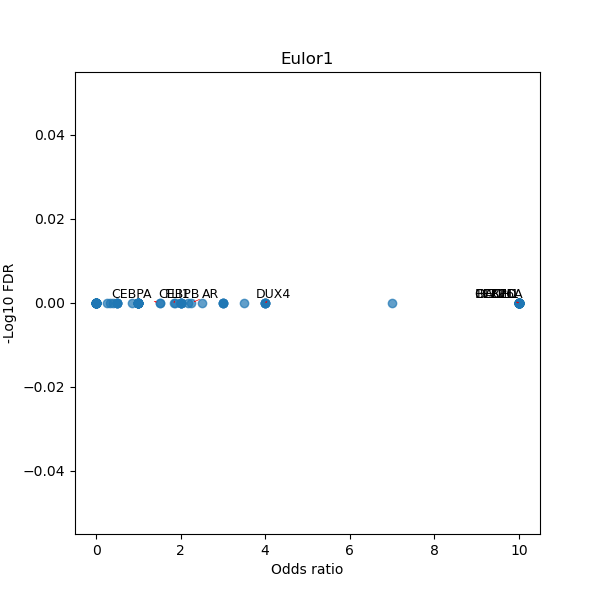
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.