EUTREP7
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001220 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Eutheria |
| Length | 392 |
| Kimura value | 27.84 |
| Tau index | 0.0000 |
| Description | Conserved repetitive element present in placental mammals. |
| Comment | 4 bp TSDs. Orientation confirmed with appropriately placed AATAAA signal. ERV1 designation is uncertain, as other LTR elements with 4 bp TSDs have been identified. |
| Sequence |
TGTAAAAGAAAAATTGTCCCAAACCTGGTCTGGGACAGAACCTTTTTGCGTGATGTAATTCACAACCTCCTTCTGGGAAGAAATTAGCATTCCTGATAAAATGCTTAAGCAATTTTCTGATAAACTTTAATGGGATTAAGACTTGTTCTGGGAAACCAGTCACCCACCCAGCCGACTGTCAGTCAAGAGTCTAGTGACCTTTAACCAGACATTCTTTTCTTTTTGTATCTAGTCTAACAATGATTATCTAACCATTCTCTTTTCCTATAAAATGTCCCTACCAAGAGNGAATCTTTGAATTGGCCTNCTAGAGGCCTTTTCCCTTGCAGCAAGNTTGNAATAAAACTTTGACACGTGCTCATAACTAAATTGTCTGAAAAGTTTCTTTAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EUTREP7 | DOF4.3 | 377 | 389 | - | 17.26 | TAAAGAAACTTTT |
| EUTREP7 | Stat5b | 146 | 154 | - | 16.36 | TTCCCAGAA |
| EUTREP7 | Stat5b | 71 | 79 | - | 16.36 | TTCCCAGAA |
| EUTREP7 | SIZF2 | 175 | 183 | - | 15.69 | ACTGACAGT |
| EUTREP7 | HEY2 | 350 | 358 | + | 15.33 | GACACGTGC |
| EUTREP7 | STAT3 | 146 | 154 | + | 15.20 | TTCTGGGAA |
| EUTREP7 | STAT3 | 71 | 79 | + | 15.20 | TTCTGGGAA |
| EUTREP7 | STAT1 | 146 | 154 | + | 15.13 | TTCTGGGAA |
| EUTREP7 | STAT1 | 71 | 79 | + | 15.13 | TTCTGGGAA |
| EUTREP7 | Stat5a::Stat5b | 146 | 154 | - | 15.12 | TTCCCAGAA |
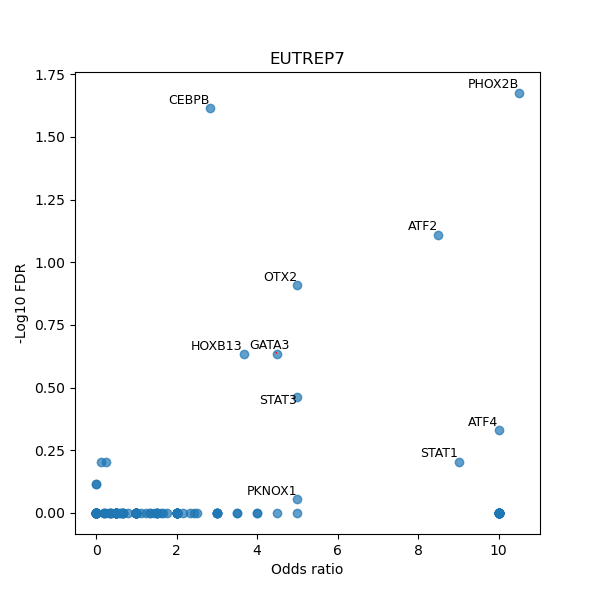
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.