EUTREP4
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001158 |
|---|---|
| TE superfamily | PiggyBac |
| TE class | DNA |
| Species | Eutheria |
| Length | 659 |
| Kimura value | 29.18 |
| Tau index | 0.0000 |
| Description | Conserved repetitive element present in placental mammals. |
| Comment | - |
| Sequence |
GTTCCCCTNCTTNCGTGTGTTTCCTTCATATTTTCTGCAGNCGTTTCAACTGAGTACTTGTTTCGGGCCTCATGGACCCAGCTATCTGGGAATAAGTTGTCATCTGNGGATAANCATTATNNNTATTTACAAGGTANAACGTTCCGTTTNGANAGAAGTNATGGCGCCTNATGGTGTCAAGAGGGGATGTGCATTCTGTTCTAGTTAAAAGNACAGGAAAACTGCCTTTCAGTGTACGTGTTGCAAGAAGCCAACGTGCACGGAGCACNATGCTTACCTTTGTGTTGATTGTGGACAGATAAATGAATAAATGATTGTCTATTTTGAGTATTAANATACAGTTGTTTCCATAGATACCTGATTATAATGACTAATTAGACAAAAGTAAGTGTTTTCCAAAGCCTATTTGAATTACANCTCTGATGTAATAAACTTCTAACTTCTTATTCAATTGGTACATGTTTCTTCTCATTTCTCTTTAAAAAAATCAATAAATNATTTGTGATGCCTTTAATAGAGATTAAATGAATAAAATCAGCTTTTACCTTATTATGAAGTTGATATTTTCAACATTTACCTTAAAAAGTAACATTGGGTCTGTCAGACCCAACGATCTGGGATTTGTTTGAAAAGTTTAATGATCTGNTTCCAGGGTTAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EUTREP4 | RFX2 | 344 | 357 | + | 13.90 | GTTTCCATAGATAC |
| EUTREP4 | DOF4.7 | 581 | 589 | - | 13.72 | TTACTTTTT |
| EUTREP4 | AG | 574 | 589 | + | 13.66 | TTACCTTAAAAAGTAA |
| EUTREP4 | RVE7L | 559 | 567 | - | 13.59 | AAAATATCA |
| EUTREP4 | ATF4 | 420 | 429 | + | 13.52 | CTGATGTAAT |
| EUTREP4 | NFATC4 | 343 | 351 | - | 13.44 | ATGGAAACA |
| EUTREP4 | mls-2 | 371 | 378 | + | 13.44 | CTAATTAG |
| EUTREP4 | mls-2 | 371 | 378 | - | 13.44 | CTAATTAG |
| EUTREP4 | ZNF24 | 305 | 317 | - | 13.42 | CAATCATTTATTC |
| EUTREP4 | HAT1 | 310 | 317 | - | 13.41 | CAATCATT |
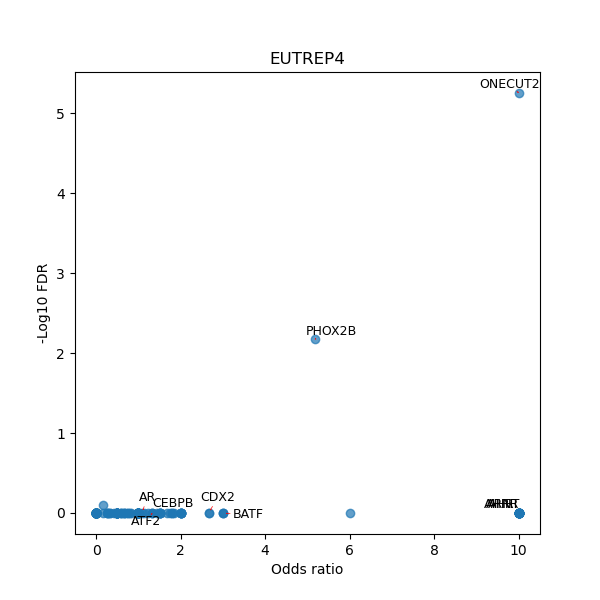
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.