EUTREP4
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001158 |
|---|---|
| TE superfamily | PiggyBac |
| TE class | DNA |
| Species | Eutheria |
| Length | 659 |
| Kimura value | 29.18 |
| Tau index | 0.0000 |
| Description | Conserved repetitive element present in placental mammals. |
| Comment | - |
| Sequence |
GTTCCCCTNCTTNCGTGTGTTTCCTTCATATTTTCTGCAGNCGTTTCAACTGAGTACTTGTTTCGGGCCTCATGGACCCAGCTATCTGGGAATAAGTTGTCATCTGNGGATAANCATTATNNNTATTTACAAGGTANAACGTTCCGTTTNGANAGAAGTNATGGCGCCTNATGGTGTCAAGAGGGGATGTGCATTCTGTTCTAGTTAAAAGNACAGGAAAACTGCCTTTCAGTGTACGTGTTGCAAGAAGCCAACGTGCACGGAGCACNATGCTTACCTTTGTGTTGATTGTGGACAGATAAATGAATAAATGATTGTCTATTTTGAGTATTAANATACAGTTGTTTCCATAGATACCTGATTATAATGACTAATTAGACAAAAGTAAGTGTTTTCCAAAGCCTATTTGAATTACANCTCTGATGTAATAAACTTCTAACTTCTTATTCAATTGGTACATGTTTCTTCTCATTTCTCTTTAAAAAAATCAATAAATNATTTGTGATGCCTTTAATAGAGATTAAATGAATAAAATCAGCTTTTACCTTATTATGAAGTTGATATTTTCAACATTTACCTTAAAAAGTAACATTGGGTCTGTCAGACCCAACGATCTGGGATTTGTTTGAAAAGTTTAATGATCTGNTTCCAGGGTTAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EUTREP4 | ceh-38 | 483 | 496 | + | 22.56 | AAAAATCAATAAAT |
| EUTREP4 | ONECUT3 | 483 | 494 | + | 19.70 | AAAAATCAATAA |
| EUTREP4 | AGL6 | 574 | 592 | + | 18.03 | TTACCTTAAAAAGTAACAT |
| EUTREP4 | AGL13 | 574 | 591 | + | 17.11 | TTACCTTAAAAAGTAACA |
| EUTREP4 | AGL15 | 574 | 589 | - | 16.03 | TTACTTTTTAAGGTAA |
| EUTREP4 | DOF4.5 | 581 | 593 | + | 15.83 | AAAAAGTAACATT |
| EUTREP4 | ONECUT1 | 485 | 493 | + | 15.50 | AAATCAATA |
| EUTREP4 | lin-54 | 404 | 416 | - | 15.44 | TGTAATTCAAATA |
| EUTREP4 | crc | 419 | 429 | + | 15.41 | TCTGATGTAAT |
| EUTREP4 | ZHD9 | 363 | 377 | + | 15.07 | TATAATGACTAATTA |
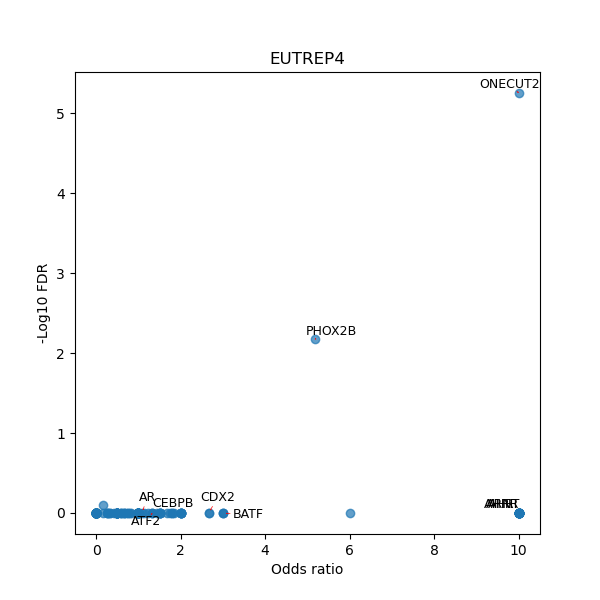
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.