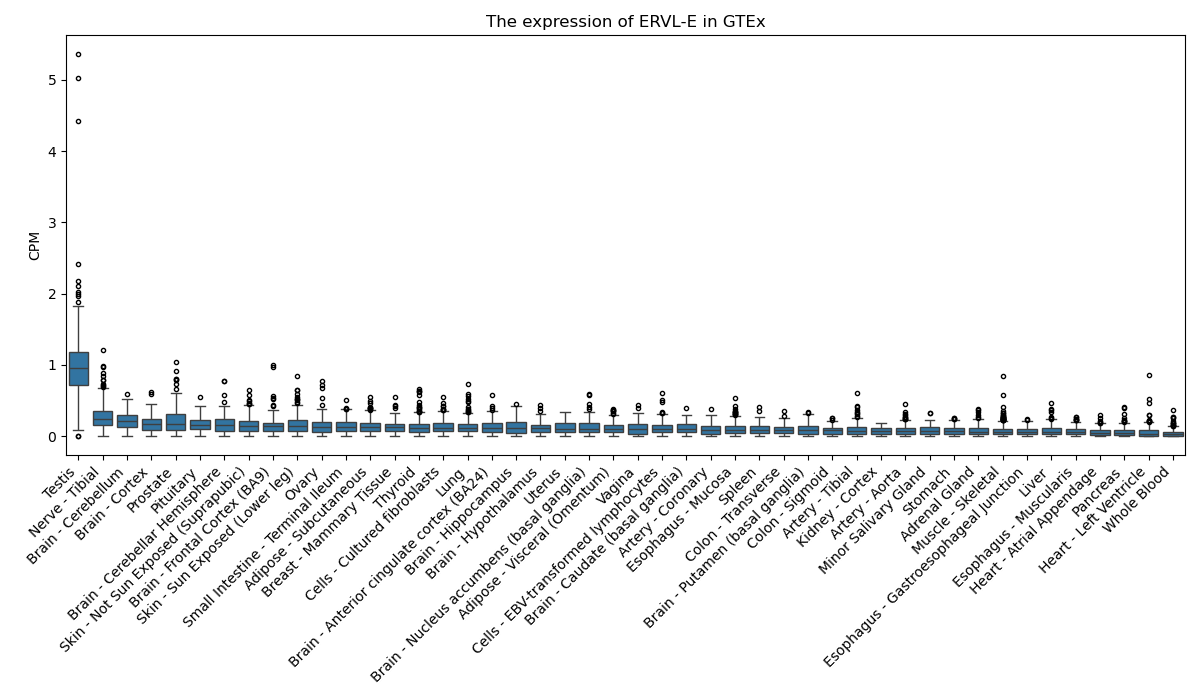
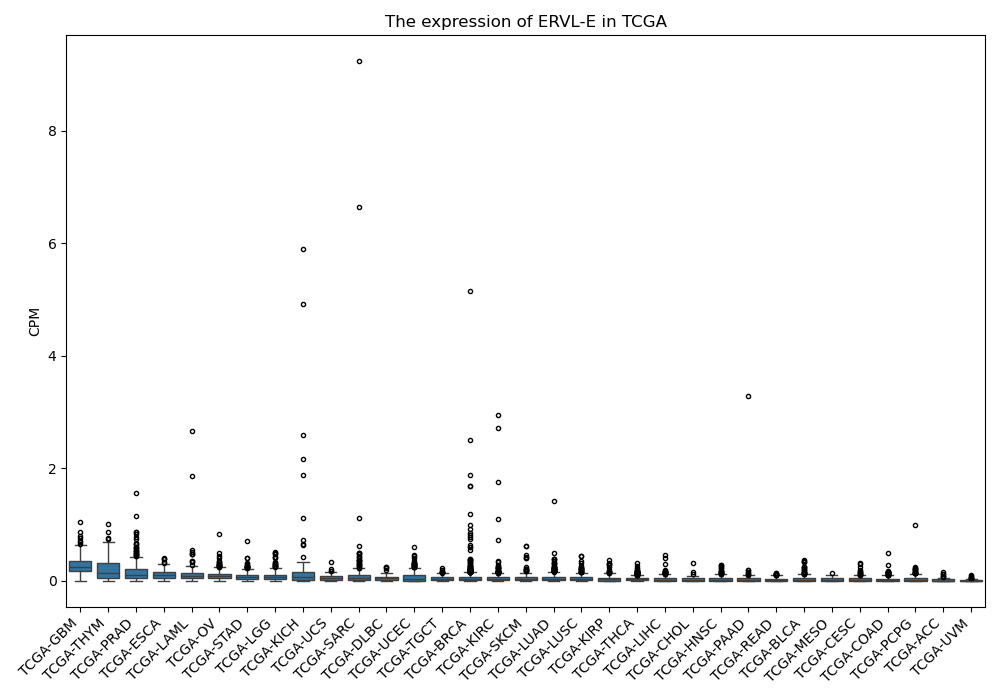
ERVL-E
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000120 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 5667 |
| Kimura value | 35.44 |
| Tau index | 0.8922 |
| Description | ERVL endogenous retrovirus, ERVL-E subfamily |
| Comment | MLT2E LTRs. ORFs at roughly pos 131-1720, 1725-5267. |
| Sequence |
ATTATTGGTACCGGGAGTGGTTCCAGGGGAACAGAACCTTAAGGATGGGAATCTGGAATTGGTTCTCTGATCTGATTAGATTTAAAGGCGCTAATGACCCTGTTTCCAGTGGTAAAGGGGACACTGGTAGTCCATGGCATGCAGTGGCAAAAAGTTACTCAAATTATCACCTGTGGNCACCTGTAATCAAGTGCCTATAGAAGGCAAGGCTTTGGGTGACCANGTANTTGCTGCCNTAGAACATTTTAGTGGAAATAAGGAGTATAATGANGTTGGTTGGTTGCTTCTAANTGCGCTGGAGAACTTGGAGAAAGAAAATGATGAGCTCAGGGCTTTAAATTCCCAGCTCAAGNTCCGGGTAAGGGACCNGAAAGCTTCTATGACTGCCCTGAAAGAAACCCTTATCTCCTGTAGCCGCAGGGCTGAGATTTCTGAAAACCAAACCCAAAGTCTNATCCTGCGGGTGGCTGAATTACAACGCAAATTGAATTCNCAACCTCGCAGGGTNTCTTNTGTTAAAGTTAGGGCATTGATTGGGAAGGAATGGGACCCTGAAAATTGGAATGGGGACATNTGGGCGGATTCCGATGAAGCTGGGGACCTTGAACCCCTAAATTCTGCCGAGCCTTCTTTGCCAGTAGAAGCAGCCCTTTCNCCCCTGTCTGAGGAGGTTAGTCTCCCCTTGCCTGAAGAANCTGTAATGGCCTCCCCTGAGGTAGTTGCCTTGCAAGGNANTGCTGATTCTCCTCAGGACCTACCCCCACCACCTCTCNTTGCTTCTAGACCTATAACTAGACTCAAGTCCCAGCAGGCCCCNAGGGGTNAGGTACAAAGTGTGACCCATGAGGAGGTANNNTACACACCAAAAGAATTGCAAGATTTTNCCAATTTATATCGACAGAAACCTGGGGAATATGTGTGGGAATGGATNCTAAGGGTGTTGGATCANGGTGGAAGGAATATAACGTTGGATCGGGCCGAATTTATTGATATGGGTNCACTAAGCAGAGATTCTGGATTCAATGTGTTAGCTCGAGNAGCTGGAAGTGGCTCTAACAGTTTGCTTGGTTGGTTGACTGAAACNTGGACTCAAAGGTGGCCTACANTNAATGAAGTTGAGATGCCAGAACTTCCTTGGTATANTGTAGAGGAAGGNATCCAAAGGCTTAGGGAGATNGGAATGTTGGAGTGGATTTATCATGTAAGACCTGCTCACCTACACCCTCTAACTATGTCCCCGGGAGGGTCCAGGGACACTCCCTTCACCAAGGCNTTGAGAAATACATTNGTGAGGGGAGCACCAGCATCCTTGAAGAGCTCTGTGGTGGCTNTTCTCTGTAGGCCAGGNATGACGGTGGGAGATGCTGCCATTGAAATGGGCTCCCTGANTTCAATGGGGATGATGGGATCCCGGGGTGGCAGAGGCCAAGTGGCGGCACTTAACCGCCAGAGACAAGGTGGGCGCGGTTACCGTAATGGGCAGCAGAGCCAAAGCGGTAATCAGAATGGTTTGACCCGCAGAGATCTTTGGCGNTGGCTAATTGATCATGGTGTCCCTAGGANTGAAATAGATGGGCAGCCTACTAAAGTCTTACTTGATTTGTATAAGCAGAAAAGCTCTAGGTCTGGTGAACAGAAGTCTGACTTGAGTCACCAACGGAGAGTCACGGCCCCTCANCCAGTTCCCAGACTTGAGCCAGTTCACAGCCAGACCCCTTGAATGAAGGGGAGGCCGGGTCCCCTTGAGGAAGGACCCTGCNACACTGCCAAAAATNTATACTGTAAATCTTCCTCCNAGCCTTCCCCAAAGGGACCTGCGGCCATTTACCAGGGTGACTGTGCACTGGGGAAAGGGAAATANCCAGACTTTTNAGGGATTACTGGACACTGGCTCTGAACTGACGCTAATTCCTGGAGACCCAAAACGCCACTGTGGTCCACCAGTCAGAGTAGGGGCTTATGGAGGTCAGGTGATNAATGGAGTTTTGGCTCGAGTCCGTCTCACAGTGGGCCCAGTGGGTCCNCGAACCCACCCTGTGGTTATTTCCCCAGTTCCNGAATGCATAGTTGGAATAGACATACTCAGCAACTGGCAGAATCCCCACATTGGTTCCCTGACCCGTGGAGTGAGGGCTATTATGGTAGGAAAGGCCAAGTGGAAGCCNCTGGAACTGCCTCTNCCTACCAAAATAGTAAACCAAAAGCAATACCGCATCCCTGGAGGAATTGCAGAGATTAGTGCCACCATCAAAGACTTGAAAGATGCAGGGGTGGTGATTCCTACCACATCCCCATTTAACTCGCCTGTTTGGCCTGTGCAGAAGACAGATGGATCTTGGAGAATGACAGTGGATTATCGTAAACTTAATCAGGTGGTGACTCCAATTGCAGCTGCTGTTCCAGATGTGGTNTCTTTACTGGAGCAAATCAACACANCCCCTGGCACCTGGTATGCAGCTATTGATCTGGCAAATGCTTTTTTCTCTATACCTGTTAGTAAAGACCACCAGAAGCAGTTTGCTTTCACCTGGCAGGGNCAGCAGTACACCTTCACTGTCTTGCCTCAGGGCTATGTCAACTCTCCNGCTCTCTGTCATAATNTAGTCCGCAGGGACCTTGATCGTCTTNNCATTCCACAGGACATCACGCTGGTCCACTACATTGATGACATCATGCTGATTGGACCTGGTGAGCAGGAAGTAGCAAGTACTCTAGACGCCTTGGTAAGACACATGCGTGCCAGAGGGTGGGAGATAAATCCCACGAAAATTCAGGGGCCTGCCACCTCGGTGAAGTTTCTAGGGGTCCAGTGGTCTGGGGCATGTCGAGATATCCCTTCCAAGGTGAAGGACAAGTTGCTGCATCTNGCNCCTCCTACCACTAAGAAAGAGGCACAATGCTTGGTGGGCCTCTTTGGATTTTGGAGGCAACATATACCNCATTTGGGCGTGCTGCTCCGACCCATTTACCGAGTAACCCGNAAGGCTGCCAGTTTTGAGTGGGGCCCAGAGCAAGAGAAGGCTCTGCAGCAGGTCCAGGCTGCNGTGCAAGCTGCTCTGCCACTTGGGCCNTATGACCCAGCAGATCCAATGGTGCTCGAAGTGTCTGTGGCAGATAGGGATGCTGTATGGAGCCTCTGGCAAGCCCCNATAGGNGAATCACAGCGCAGACCCCTAGGATTTTGGAGCAAAGCCATGCCATCTTCTGCAGATAACTATTCTCCTTTTGAGAAACAGCTCCTGGCTTGCTACTGGGCCCTGGTAGAGACTGAACGCCTGACCATGGGNCACCAAGTNACCATGCGACCTGAGCTGCCCATCATGAACTGGGTGTTATCTGACCCACCAAGCCATAAAGTTGGGCGTGCACAGCAGCANTCCATCATCAAGTGGAAGTGGTATATACGAGATCGGGCTCGAGCAGGTCCNGAAGGCACAAGTAAGTTGCATGAGCAGGTGGCTCAGACTCCCATGGCNCCTACTCCTGCTGCATTGCCTCCTCTCCCTCAACCCGCACCTATGGCCTCATGGGGAGTTCCCTATGACCAGTTGACTGAGGAAGAAAAAACTCGGGCCTGGTTTACAGATGGTTCTGCACGATATGCTGGCACCANCCGAAAGTGGACGGCTGCAGCACTACAGCCCCACTCAGGGGTGGCCCTGAAGGACAGTGGTGAAGGGAAATCCTCCCAGTGGGCAGAACTTCGAGCAGTGCACCTGGTTGTCCACTTTGCCTGGAAGGAGAGATGGCCAGAGGTACGGATCTACACTGATTCATGGGCAGTGGCTAACGGTTTGGCTGGATGGTCAGGGACTTGGAAGGAACANGATTGGAAGATTGGTGACAAGGAGGTCTGGGGAAGAGGTATGTGGATGGACCTCTCGGAATGGGCACAGAGTGTGAAGATATTTGTGTCCCATGTGAATGCTCACCAAAGGGCANCCNCNGCAGAGGAGGNTCTCAATAATCAGGTGGACAAGATGACCCGTTCTGTGGATGTCAGTCAGCCTCTTTCCCCAGCCACCCCNGTGCTTGCTCAATGGGCTCATGAACAAAGTGGCCATGGTGGCAGGGATGGAGGCTATGCATGGGCTCAGCAACATGGACTTCCNCTCACCAAGGCTGATCTGGCTACNGCCACTGCTGAGTGCCCAACCTGCCAACAGCAGAGACCAACGCTGAGCCCCCGATATGGCACCATTCCCCGGGGGGACCAGCCAGCCACCTGGTGGCAGGTTGATTACATTGGACCNCTTCCATCATGGAAGGGGCAGCGATTTGTCCTCACTGGAATAGACACTTATTCTGGATATGGATTTGCCTTCCCTGCCCGCAATGCTTCTGCCAGNACCACCATCCGTGGACTTACAGAATGCCTTATTCACCGTCATGGTATTCCACACAGCATTGCTTCTGACCAAGGAACTCATTTTACAGCAAANGAAGTGCGGCAATGGGCTCATGCCCATGGAATTCACTGGTCTTACCACGTNCCCCATCACCCNGAAGCAGCTGGCCTGATAGAACGGTGGAATGGCCTNTTGAAGACTCAGTTACGGCGCCAGCTGGGNGACAACACCTTGNAGGGCTGGGGTNNTGTCCTNCAGGATGCGGTATATGCTCTGAATCAGCGACCAATATATGGTGCTGTTTCTCCCATAGCCAGAATNCACGGGTCCGGGAATCAAGGGGTGGAAGTGGGAGTGGCTCCTCTCACTATTACNCCTAATGACCCACTNGCAAAATTTTTGCTTCCCGTCCCCGCAACTTTGGGCTCTGCTGGTTTAGAGGTCTTAGTTCCCAAGGGAGGAATGCTTCCACCAGGGGACACAACAATGGTTCCATTGAACTGGAAGCTGAGACTGCCACCTGGCCACTTTGGGCTCCTCATGCCACTGAACCAACAGGCAAAGAAGGGAGTTACTGTACTGGCTGGGGTGATTGATCCTGATTATCAAGGGGAAATTGGGTTGCTGCTACACAATGGGGGCAAGGAGGANTATGTCTGGAATNCAGGAGATCCTCTGGGGCGCCTCTTAGTACTCCCATGTCCNGTGATAAAAGTTAATGGAAAACTACAGCAACCCAATANAGGCAGGACCGCTAANGGCTCAGACCCTTCAGGAATGAAGGTTTGGGTCACCCCACCAGGCAAAGAACCACGACCAGCTGAGGTGCTNGCTGAGGGCAAAGGGAATATGGAATGGGTAGTGGAAGAAGGAAGTTATAAATACCAGCTACGACCNCGTGACCAGTTGCAGAAACGAGGACTGTAGTAGTTATGNGTATTTCTTCCTTGCTTTGATATGAATATATTTGTGATATATATATTAACNAATATCTTTNTTTTCTTTCCTCTCTCATTCCCCTACTATCTAACATAAGATGTGTTAATAGTAGTTAACCTTATATCTCAGTATTTAAGTTACAGGATATCAAAGGGGGANTGTGACTCAGCTAGAAGAGNAATGAACATCACCCAGAGATGGATAAAGTGACNTNTGGGACTTTGTATCCTCTTTTGGGGAGAGGGTTAGCGTGTTTTCGGTTGTACGAGGGATAGTTGCATCATGTTAGGCGGAAGCATGATTTTGCTATTGTCTTTATTTGGAAGTTAAATATGGTTNAAAGAGGTGTGTATGGATGCCGAGTTGACAAGGGGTGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERVL-E | Ebf2 | 4800 | 4808 | + | 16.79 | CCCAAGGGA |
| ERVL-E | EBF3 | 4800 | 4808 | + | 16.59 | CCCAAGGGA |
| ERVL-E | CG7928 | 2211 | 2222 | + | 16.57 | CGCATCCCTGGA |
| ERVL-E | TCP1 | 2999 | 3009 | - | 16.57 | CTGGGCCCCAC |
| ERVL-E | odd | 3245 | 3255 | - | 16.53 | CCCAGTAGCAA |
| ERVL-E | CTCF | 4221 | 4235 | + | 16.49 | GCCACCTGGTGGCAG |
| ERVL-E | MGA::EVX1 | 162 | 172 | - | 16.49 | AGGTGATAATT |
| ERVL-E | SPT15 | 5321 | 5331 | + | 16.40 | GATATATATAT |
| ERVL-E | ZNF211 | 3395 | 3404 | - | 16.39 | TATATACCAC |
| ERVL-E | HOXD12::ELK1 | 5219 | 5231 | + | 16.39 | AGGAAGTTATAAA |
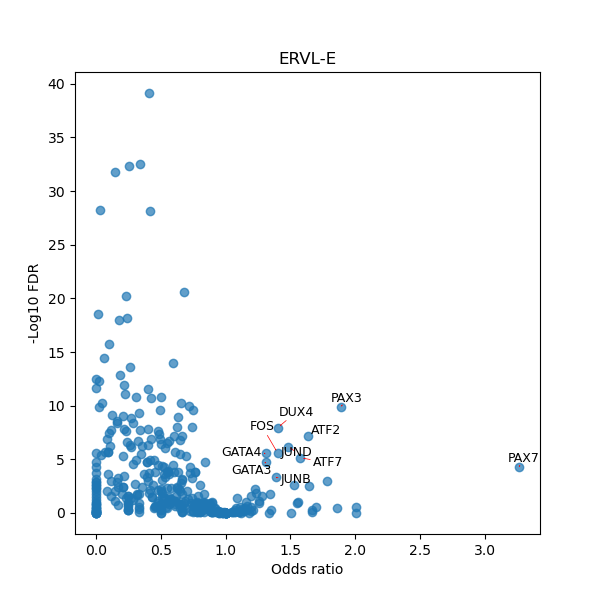
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.