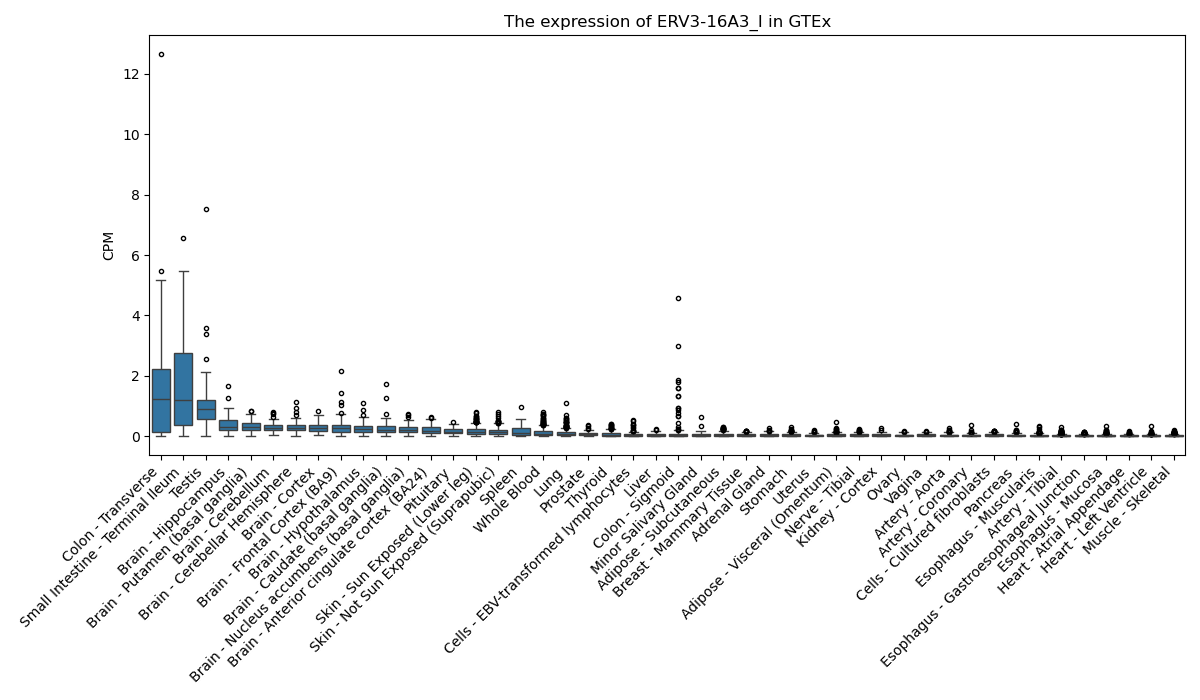
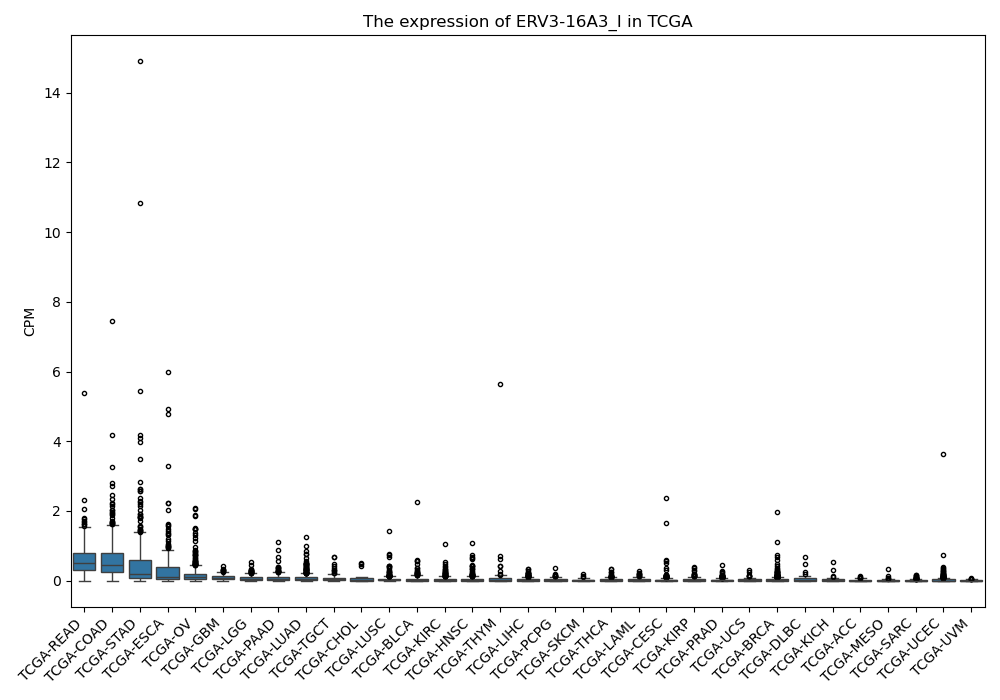
ERV3-16A3_I
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000116 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Theria_mammals |
| Length | 5226 |
| Kimura value | 31.08 |
| Tau index | 0.8928 |
| Description | Internal region of an ERVL endogenous retrovirus, ERV3-16A3_I subfamily |
| Comment | Reconstructed from the human genome, but it's shared by all placental mammals. |
| Sequence |
CATGAGTGGTACCGGGAGTGGTCCGNAGAAAACAGGCGGTAAGATGGGGTTTTGGGACTGGCTCACTCACCGCCCGGCGGACAGAGGATACCATCCTGGCACAAGGTGGCGGCCCAGTTGCTAAAGATTTCACCGGTGGTGACCTGGGAAAATGTGTCCCAGNNNTTGGAAGGGGAATGCCCTTGCAGGTGCAATGATTCAGGCATTTGAAAGATATGGGGAGANAATGCCTACAAGGACAGCGGAGTTGGCTGGTTATTGCTAAGTTGTATTGATGCCCTGCAGAGGGATAATGAGAAACTGAGGGCNGTTAACAAGCAGTTAAAGGCTAAGTGTGAGAGCCAGAGGGCCTCTTTGNTGGTAGCTTACAAAGAGGCCCTTATCTCCTGCAGTGGAAGAGCAGACANAGCTGAGGAGCAGACTCAGGATCTAATAGTTAGAGTCGCAGAGCTCCAGAGACGTTTGAATGCTCAGCCAAGGCGAGGTCTGTTATGCTAAAGNTAAGGCCCTTGGTTGGGAAAACCTGGGACGGAACACGGGATGGGGACATCTGGGTGGATGCCCCCGAAGATNTTGGCTCTCCAGACTCCTCTGAACCCTCNGAGCCTGCAGAAGTGGCCCACCCCTCCCTANTAAGAGCTAGCACTCCTCTTGCGCCCGCGCTGGAAGACGNTGCAGAGGCCTCTCCCCGCAAGGCAACAGGTGCCCCCTCTCAGGANCTGCCCCCACCTCCCCTCCTGGCCGCCAGGCCGATAACTAGGGTTAAGTCNCAGCATAACCCGGCTGGGGACGTGCTGGGCCTGATAAGGGAGGAAAGGGACTATACCCCAAAGGAGCTGCAAGAATTAGCCAGCATGTACCGGCAGGAGCCAGGGGAGTATACNCNTGGGACTGGATTTTGAGGGTGCTTGATCAAGGGGGCCGGAATATAAGCGGTAAGACTGGATAANGGAAGAATTTATTGACTTGGGAGCACTTTCTCGGGATACAGGATTTAACACCCTGGCAAGGACCCCAGGAGATGGTGCAAACTCGCTGCTAGGATGGCTCCTAGAAGCGAGAAAAATGGCCCACGCTGAGCGAAGTNGAAATGCCNGAATTGCCNTGGCAGACGGTAGAGGAAGGGATTAAAGGCTCAGGGAAGTGGGCATGCTGGAATGGATATNATTATGTAAGGCCGGAAGACCCACCAGANGATTATGTTCCACGGGAGGGCCCACACCATTCACCAAGGCCATNAGGAATGCGCTGGTGAGAGGGGCACCAGCANNTCACTAAGAAGTTCAGTGGTGGCTCTCCTCTGCAGGCCAGGGCTGACGGTAGGAGAGGCCGTTACAGAACTGGGCTCGCTGATAGCAATGGGGATGATAGGACCCCGAAACAATAGAGGCCAGGTGGCGGCGCTTAACCGCCAGAAGCCAGGNGGNCGCAATTATCGTAATGACCGGAAGGTCGGAGTGGCAGCCAAGGGGGCCTGACCCAAGAGAGTTGTGGAGATGGTTAATAGAACACGGCGTCCCTAGGGGCAAAATAGATNGACAAGGGTGCTGCTTAACTTGTAACAGCAGAAAAAATCTGGGAGGCTGAGGGCGGTCGCCCCAATAAAAAAGTCACGATCCCTTGCCCAGTTTCCGGACCTGAGCCAGTTTTCAGACCCGGAACCCATTGACTGAAGAGGNGGCCGGGTCCCCAGGAGGAAGGACACTGCAATGATTCTTCTCNTTCCTCCCGNAGGGACCTATGGCCATTTACTCGGGTAACTGTACACTGGGGAAAGGGAATACCCAGACATTTCGAGGACTGTTGGACACAGGGTCTGAGTTGACATTGATACCCGGAGACCCGAAGCGTCATCATGGCCCCCTGTTAGAGTGGGGGCATATGGGGGCCAGGTAATAAATGGAGTCCTGGCCAAGGTCCGGCTCACAGTGGGTCCACTGGGTCCANGAACCCAGTGGTCATTTCCCCGGTCCCCGAATGTATAATTGGGTAGTTGGAGTAACCCCCACATTGGNTCCTTGGCCTGTGGGGTAAGAGCTATCATAGTGGGGAAGGCCAAGTGGAAGCCTCTGAAACTGCCCNCCNCCNCCATTCCTAGCCAAGATAGTAAATTATTATATCCTCTCGAGGAAGTGGCGGATGCAGATTAGTGCCACCNTTAAAGACCTAAAGGATGCAGGGGTGGTGGTCCCCATCATATCTCCATTTAATTCACCAGTCTGGCCCCTGCAGAAACCGGATGGATCCTGGAGAATGACNGTAGACTACCGCAAACTCAACCAAGTAGTAGCCCCGATTGCAGCTGCTGTGCCAGATGTGGTATCTTTGCTAGAGCAGATTAACACGGCCTCAGGTACATGGTATGCGGCCATTGATTTGGCGAATGCGTTCTTTTCCATTCCNATCAGAAAAGAGGATCAGAAACATTCACATGAACAGGAAANAATATACATTTACAGTTTTGCCCCAGGGCTATGTTATCTTCTCCTCTGTCATAATAAGTCCGAAGAGATCTGGACCGTCTGGACATCCCGCAGAACATCACATTGATCCATTACATCGATGACATCATGCTGATCGGACNGGATGAGCAAGAGGTGGCTACGTTAGGCCTTGGTAAGACACATGCGCTGAGGGTGGAGATAACCCTACGAAGATTCAGCGNNNGGACCTGCCACNTCAGTAAAGTTTTTAGGGGTCCAGTGGTCNGGGGCATGCCGGGACATCCCCTCCAAAGTAAAAGACAAATTGTTGCATCTTGCATCTCCTACCACGAAGAAGGAAGCACAACGCCTGGTAGGCCTCTTTGGGTTCTGGAGGCAGCATATTCCACACCTGGGAATACTGCTCCGGCCCATNTACCGGGTGACACGGAAGGCTGCCAGCTTTGAGTGGGCCCAGAGCAGGAAAGGGCTCTGCAGCAGGTCCAGGCTGCGGTGCAAGCAGCCCTGCCGCTTGGGCCATACGACCCGGCAGACCCTATGGTGTTGGAGGTGTCAGTGGTGGGAAAAGATGCNGTGTGGAGTTTATGGCAAGCCCCAGTGGGAGAATCACAACGCAGGCCCCTGGGGTTCTGGAGCAAGGCTGCAAGTCTTTTGAAAAACAGCTCTTNNNATGCTACTGGTAGAGANGGAACGCTTACCATGGGACACCAAGTGACACGNANCAAAATGTACCCCGTCATGAGCTGGGTTCTGTCGGACCCACCAAGTCATAAGGTCGGGCGGGCCCAGCAGCAATCCATCGTAAGATGGAAGTGGTACATCCGGGATCGAGCACGAGGAGGACCACAAGTAAGCTGCATGAGCAGGTAGCCCAGACCCCCATGTCACCCACCACNGTTGCACCAGCGCTCCTTCAGCTCACACCTATGGCCGTATGGAGNAGAGGTGTGNTCCTGATCGACCAGCTGAAGGAGGAGGAAAAANGCCCGAGCTTGGTTCACGGATGGGTCGGCTCGGTATGTGGGTGCAAGCCGAAAATGGACGGCGGCTTGCACTACAGCCNCACTCAGGGGTGGCCTTGAAAGACAGTGGNGAGGGAAAATCTTCCCAATGGGCAGAGCTTCGGGCGGTGCACCTGGTCATCCACTTTGTGTGGAAGGAGAAGTGGCCCGAGGTNAGAATATATACGGACTCATGGGCAGTGGCGAATGGCTTGGCCGGCTGGTCAGGGGCCTGGAAGGAGAAAGATTGGAAGATCGGAGACAAGGAGGTCTGGGGANTAGAGGCATGTGGATGGACNTATGGGAGTGGGCACGAAGTGTGAAGATCTTTGTATCACATGTTAACGCCCACCAGAGAGCATCCACCACGGAAGAGGCACTAAACAACCAAGTAGACAAAATGACTCGGCCAGTTGACGTCAGCCAGCCTCTGTCATCGGCCACCCCANGTGCTGGCACAATGGGCACATGAACGGAGTGGCCACGGTGGCAGAGATGGAGGCTACGCATGGGCCCAACAGCATGGACTCCCACTCACCAAGGCTGATCTAGCTACTTGCTGCCNCNNCTGAATGTCCAACCTGCCAGCAACAGAGACCGATCCCCGATATGGCACCATTCCTCGAGGAGACCAACCAGCCACTTGGTGGCAAGTTGACTACATTGGGCCCCTTCCATCCTGGAAGGGNCAGCGGTTCATCCTNACAGGAATACATATTCCGGGTATGGGTTTGCCTTTCCTGCCCGCAGGGCCTCAGCCAGCACCACTATCCGAGGGCTTACGGAGTGCCTGATCCACNGGCATGGGATCCCACACAACATNGCATCNGACCAGGGACCCACTTTACAGCAAAGGAGGTGCGGGAGTGGGCCCATGACCATGGGATCCACTGGTCNTATCACATACCGCACCATCCAGAAGCTGCCGGCCTGATAGAGCGNTGGAACGGCCTNCTGAAGGCACAGCTGAAGCGCCAGCTCGGAGACGATACTCTGCGAGGATGGGGCGCCATCCTTCAGGATGCAGTGTCCCCAATAGGAAGAATACATGGGTCCGGGAACCAAGGGGTGGAAGCAGGAGTGTNGCCCCACTTACCATCACTCCCAGTGACCCACTGGGGGANTTTGTGCTTCCCGTCCCCGCAACTCTGGGCTCTGCAGGNTTAGAGGTCCTGGTCCCCAAAGGGGGNACGCTCTCGCCAGGGGACACAGCGTCCCATTGAACTNTANAGCTACGGCTGCCGCCTGGGCACTTTGGGCTCCTTGTGTCCAGGGACCAGCAGGCAAGAAGAGGAGTCACCATCTTGGCAGGGGTAATTGACCCTGATCATCAGGAGGAGGTAGGGCTGCTNTTACACAATGGGGGCAGGGAGGAATACGTNTGGAACCCAGGTGATCCACTTGGGCGCCTCTTGGTACTCCCTTGCCCAATTNTGACTGTAAATGGACAAGTGCAGCAACCCCGGCCTGAGAAGGGCATGGTNACCAGGGGCTCAGACCCCTCAGGAATGAGGGTCTGGGTCACGCCACCAGGTAAGCCACCGAGACCAGCAGAGTGNTAGCTGAGGGTGAGGGGAATCTAGAATGGATAGTGGAGGAGGGAGACGATGAGTATCAGTTGCGGCCCCGAGACCAACTGAGCGACGGGGGCTGTAGTTCGTCCCACTAACCTCCCTCTTCTAAGTTTCCCTCAGGAAGAGAGGCCCACNGGAATCCTGGAGGAGCTGCTCCTGAACCGAACGTGTATGGAGAAGTGGATCCGNGCGGCGCAAGGGGTGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERV3-16A3_I | IDD6 | 2735 | 2750 | - | 16.54 | AATTTGTCTTTTACTT |
| ERV3-16A3_I | IDD6 | 2735 | 2750 | - | 16.53 | AATTTGTCTTTTACTT |
| ERV3-16A3_I | ZNF701 | 4502 | 4518 | + | 16.52 | GGGAACCAAGGGGTGGA |
| ERV3-16A3_I | ARALYDRAFT_897773 | 2185 | 2193 | - | 16.39 | GGGGACCAC |
| ERV3-16A3_I | Prdm15 | 518 | 528 | + | 16.38 | GAAAACCTGGG |
| ERV3-16A3_I | TCP23 | 1213 | 1220 | + | 16.36 | GGGCCCAC |
| ERV3-16A3_I | TCP23 | 2892 | 2899 | - | 16.36 | GGGCCCAC |
| ERV3-16A3_I | TCP23 | 4312 | 4319 | - | 16.36 | GGGCCCAC |
| ERV3-16A3_I | ERF076 | 1395 | 1404 | - | 16.36 | GCGCCGCCAC |
| ERV3-16A3_I | TFAP2C | 4187 | 4199 | - | 16.34 | GGCCCTGCGGGCA |
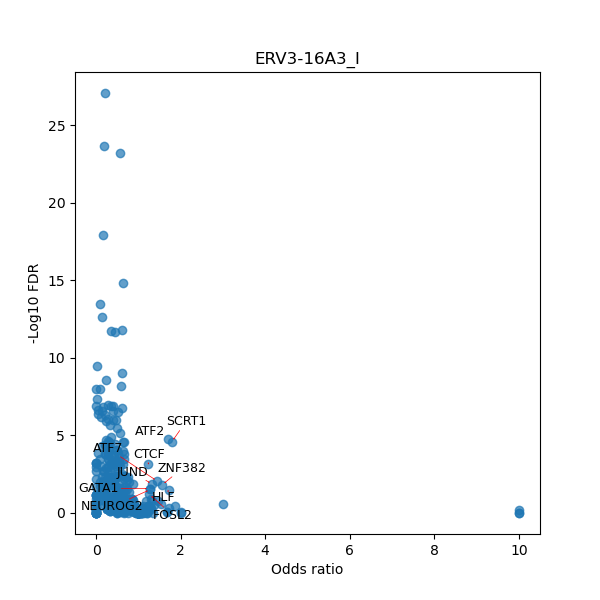
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.