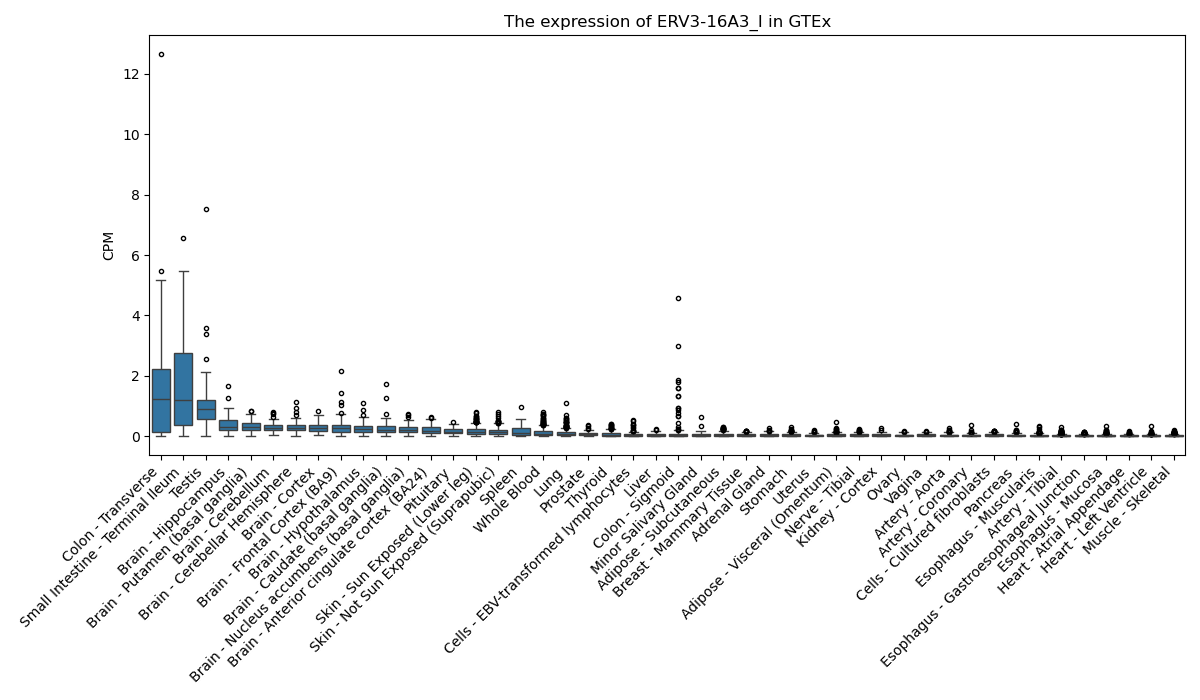
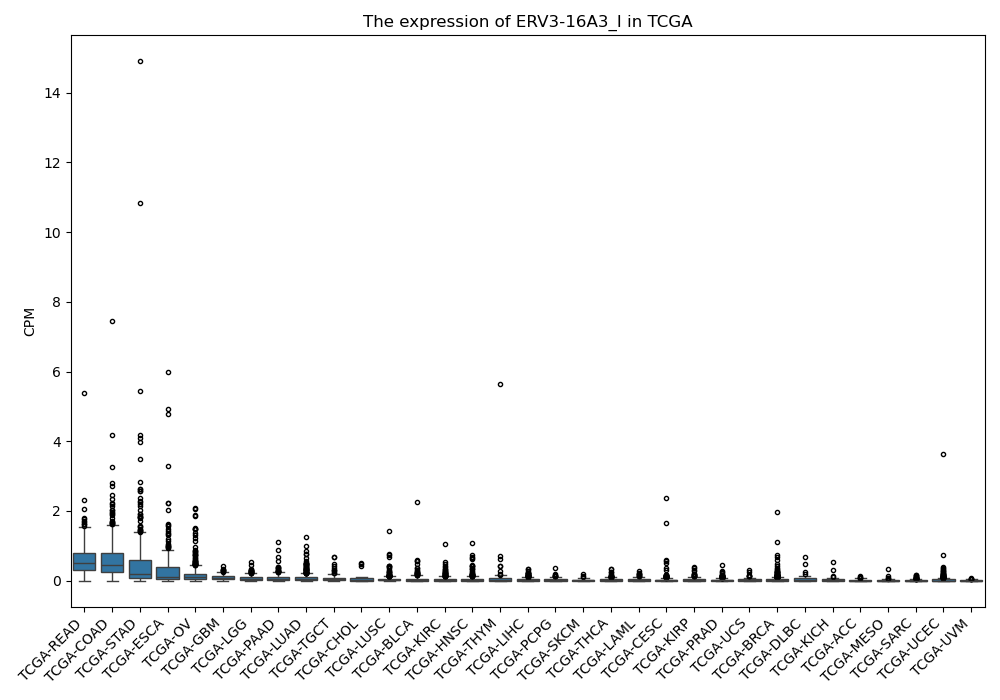
ERV3-16A3_I
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000116 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Theria_mammals |
| Length | 5226 |
| Kimura value | 31.08 |
| Tau index | 0.8928 |
| Description | Internal region of an ERVL endogenous retrovirus, ERV3-16A3_I subfamily |
| Comment | Reconstructed from the human genome, but it's shared by all placental mammals. |
| Sequence |
CATGAGTGGTACCGGGAGTGGTCCGNAGAAAACAGGCGGTAAGATGGGGTTTTGGGACTGGCTCACTCACCGCCCGGCGGACAGAGGATACCATCCTGGCACAAGGTGGCGGCCCAGTTGCTAAAGATTTCACCGGTGGTGACCTGGGAAAATGTGTCCCAGNNNTTGGAAGGGGAATGCCCTTGCAGGTGCAATGATTCAGGCATTTGAAAGATATGGGGAGANAATGCCTACAAGGACAGCGGAGTTGGCTGGTTATTGCTAAGTTGTATTGATGCCCTGCAGAGGGATAATGAGAAACTGAGGGCNGTTAACAAGCAGTTAAAGGCTAAGTGTGAGAGCCAGAGGGCCTCTTTGNTGGTAGCTTACAAAGAGGCCCTTATCTCCTGCAGTGGAAGAGCAGACANAGCTGAGGAGCAGACTCAGGATCTAATAGTTAGAGTCGCAGAGCTCCAGAGACGTTTGAATGCTCAGCCAAGGCGAGGTCTGTTATGCTAAAGNTAAGGCCCTTGGTTGGGAAAACCTGGGACGGAACACGGGATGGGGACATCTGGGTGGATGCCCCCGAAGATNTTGGCTCTCCAGACTCCTCTGAACCCTCNGAGCCTGCAGAAGTGGCCCACCCCTCCCTANTAAGAGCTAGCACTCCTCTTGCGCCCGCGCTGGAAGACGNTGCAGAGGCCTCTCCCCGCAAGGCAACAGGTGCCCCCTCTCAGGANCTGCCCCCACCTCCCCTCCTGGCCGCCAGGCCGATAACTAGGGTTAAGTCNCAGCATAACCCGGCTGGGGACGTGCTGGGCCTGATAAGGGAGGAAAGGGACTATACCCCAAAGGAGCTGCAAGAATTAGCCAGCATGTACCGGCAGGAGCCAGGGGAGTATACNCNTGGGACTGGATTTTGAGGGTGCTTGATCAAGGGGGCCGGAATATAAGCGGTAAGACTGGATAANGGAAGAATTTATTGACTTGGGAGCACTTTCTCGGGATACAGGATTTAACACCCTGGCAAGGACCCCAGGAGATGGTGCAAACTCGCTGCTAGGATGGCTCCTAGAAGCGAGAAAAATGGCCCACGCTGAGCGAAGTNGAAATGCCNGAATTGCCNTGGCAGACGGTAGAGGAAGGGATTAAAGGCTCAGGGAAGTGGGCATGCTGGAATGGATATNATTATGTAAGGCCGGAAGACCCACCAGANGATTATGTTCCACGGGAGGGCCCACACCATTCACCAAGGCCATNAGGAATGCGCTGGTGAGAGGGGCACCAGCANNTCACTAAGAAGTTCAGTGGTGGCTCTCCTCTGCAGGCCAGGGCTGACGGTAGGAGAGGCCGTTACAGAACTGGGCTCGCTGATAGCAATGGGGATGATAGGACCCCGAAACAATAGAGGCCAGGTGGCGGCGCTTAACCGCCAGAAGCCAGGNGGNCGCAATTATCGTAATGACCGGAAGGTCGGAGTGGCAGCCAAGGGGGCCTGACCCAAGAGAGTTGTGGAGATGGTTAATAGAACACGGCGTCCCTAGGGGCAAAATAGATNGACAAGGGTGCTGCTTAACTTGTAACAGCAGAAAAAATCTGGGAGGCTGAGGGCGGTCGCCCCAATAAAAAAGTCACGATCCCTTGCCCAGTTTCCGGACCTGAGCCAGTTTTCAGACCCGGAACCCATTGACTGAAGAGGNGGCCGGGTCCCCAGGAGGAAGGACACTGCAATGATTCTTCTCNTTCCTCCCGNAGGGACCTATGGCCATTTACTCGGGTAACTGTACACTGGGGAAAGGGAATACCCAGACATTTCGAGGACTGTTGGACACAGGGTCTGAGTTGACATTGATACCCGGAGACCCGAAGCGTCATCATGGCCCCCTGTTAGAGTGGGGGCATATGGGGGCCAGGTAATAAATGGAGTCCTGGCCAAGGTCCGGCTCACAGTGGGTCCACTGGGTCCANGAACCCAGTGGTCATTTCCCCGGTCCCCGAATGTATAATTGGGTAGTTGGAGTAACCCCCACATTGGNTCCTTGGCCTGTGGGGTAAGAGCTATCATAGTGGGGAAGGCCAAGTGGAAGCCTCTGAAACTGCCCNCCNCCNCCATTCCTAGCCAAGATAGTAAATTATTATATCCTCTCGAGGAAGTGGCGGATGCAGATTAGTGCCACCNTTAAAGACCTAAAGGATGCAGGGGTGGTGGTCCCCATCATATCTCCATTTAATTCACCAGTCTGGCCCCTGCAGAAACCGGATGGATCCTGGAGAATGACNGTAGACTACCGCAAACTCAACCAAGTAGTAGCCCCGATTGCAGCTGCTGTGCCAGATGTGGTATCTTTGCTAGAGCAGATTAACACGGCCTCAGGTACATGGTATGCGGCCATTGATTTGGCGAATGCGTTCTTTTCCATTCCNATCAGAAAAGAGGATCAGAAACATTCACATGAACAGGAAANAATATACATTTACAGTTTTGCCCCAGGGCTATGTTATCTTCTCCTCTGTCATAATAAGTCCGAAGAGATCTGGACCGTCTGGACATCCCGCAGAACATCACATTGATCCATTACATCGATGACATCATGCTGATCGGACNGGATGAGCAAGAGGTGGCTACGTTAGGCCTTGGTAAGACACATGCGCTGAGGGTGGAGATAACCCTACGAAGATTCAGCGNNNGGACCTGCCACNTCAGTAAAGTTTTTAGGGGTCCAGTGGTCNGGGGCATGCCGGGACATCCCCTCCAAAGTAAAAGACAAATTGTTGCATCTTGCATCTCCTACCACGAAGAAGGAAGCACAACGCCTGGTAGGCCTCTTTGGGTTCTGGAGGCAGCATATTCCACACCTGGGAATACTGCTCCGGCCCATNTACCGGGTGACACGGAAGGCTGCCAGCTTTGAGTGGGCCCAGAGCAGGAAAGGGCTCTGCAGCAGGTCCAGGCTGCGGTGCAAGCAGCCCTGCCGCTTGGGCCATACGACCCGGCAGACCCTATGGTGTTGGAGGTGTCAGTGGTGGGAAAAGATGCNGTGTGGAGTTTATGGCAAGCCCCAGTGGGAGAATCACAACGCAGGCCCCTGGGGTTCTGGAGCAAGGCTGCAAGTCTTTTGAAAAACAGCTCTTNNNATGCTACTGGTAGAGANGGAACGCTTACCATGGGACACCAAGTGACACGNANCAAAATGTACCCCGTCATGAGCTGGGTTCTGTCGGACCCACCAAGTCATAAGGTCGGGCGGGCCCAGCAGCAATCCATCGTAAGATGGAAGTGGTACATCCGGGATCGAGCACGAGGAGGACCACAAGTAAGCTGCATGAGCAGGTAGCCCAGACCCCCATGTCACCCACCACNGTTGCACCAGCGCTCCTTCAGCTCACACCTATGGCCGTATGGAGNAGAGGTGTGNTCCTGATCGACCAGCTGAAGGAGGAGGAAAAANGCCCGAGCTTGGTTCACGGATGGGTCGGCTCGGTATGTGGGTGCAAGCCGAAAATGGACGGCGGCTTGCACTACAGCCNCACTCAGGGGTGGCCTTGAAAGACAGTGGNGAGGGAAAATCTTCCCAATGGGCAGAGCTTCGGGCGGTGCACCTGGTCATCCACTTTGTGTGGAAGGAGAAGTGGCCCGAGGTNAGAATATATACGGACTCATGGGCAGTGGCGAATGGCTTGGCCGGCTGGTCAGGGGCCTGGAAGGAGAAAGATTGGAAGATCGGAGACAAGGAGGTCTGGGGANTAGAGGCATGTGGATGGACNTATGGGAGTGGGCACGAAGTGTGAAGATCTTTGTATCACATGTTAACGCCCACCAGAGAGCATCCACCACGGAAGAGGCACTAAACAACCAAGTAGACAAAATGACTCGGCCAGTTGACGTCAGCCAGCCTCTGTCATCGGCCACCCCANGTGCTGGCACAATGGGCACATGAACGGAGTGGCCACGGTGGCAGAGATGGAGGCTACGCATGGGCCCAACAGCATGGACTCCCACTCACCAAGGCTGATCTAGCTACTTGCTGCCNCNNCTGAATGTCCAACCTGCCAGCAACAGAGACCGATCCCCGATATGGCACCATTCCTCGAGGAGACCAACCAGCCACTTGGTGGCAAGTTGACTACATTGGGCCCCTTCCATCCTGGAAGGGNCAGCGGTTCATCCTNACAGGAATACATATTCCGGGTATGGGTTTGCCTTTCCTGCCCGCAGGGCCTCAGCCAGCACCACTATCCGAGGGCTTACGGAGTGCCTGATCCACNGGCATGGGATCCCACACAACATNGCATCNGACCAGGGACCCACTTTACAGCAAAGGAGGTGCGGGAGTGGGCCCATGACCATGGGATCCACTGGTCNTATCACATACCGCACCATCCAGAAGCTGCCGGCCTGATAGAGCGNTGGAACGGCCTNCTGAAGGCACAGCTGAAGCGCCAGCTCGGAGACGATACTCTGCGAGGATGGGGCGCCATCCTTCAGGATGCAGTGTCCCCAATAGGAAGAATACATGGGTCCGGGAACCAAGGGGTGGAAGCAGGAGTGTNGCCCCACTTACCATCACTCCCAGTGACCCACTGGGGGANTTTGTGCTTCCCGTCCCCGCAACTCTGGGCTCTGCAGGNTTAGAGGTCCTGGTCCCCAAAGGGGGNACGCTCTCGCCAGGGGACACAGCGTCCCATTGAACTNTANAGCTACGGCTGCCGCCTGGGCACTTTGGGCTCCTTGTGTCCAGGGACCAGCAGGCAAGAAGAGGAGTCACCATCTTGGCAGGGGTAATTGACCCTGATCATCAGGAGGAGGTAGGGCTGCTNTTACACAATGGGGGCAGGGAGGAATACGTNTGGAACCCAGGTGATCCACTTGGGCGCCTCTTGGTACTCCCTTGCCCAATTNTGACTGTAAATGGACAAGTGCAGCAACCCCGGCCTGAGAAGGGCATGGTNACCAGGGGCTCAGACCCCTCAGGAATGAGGGTCTGGGTCACGCCACCAGGTAAGCCACCGAGACCAGCAGAGTGNTAGCTGAGGGTGAGGGGAATCTAGAATGGATAGTGGAGGAGGGAGACGATGAGTATCAGTTGCGGCCCCGAGACCAACTGAGCGACGGGGGCTGTAGTTCGTCCCACTAACCTCCCTCTTCTAAGTTTCCCTCAGGAAGAGAGGCCCACNGGAATCCTGGAGGAGCTGCTCCTGAACCGAACGTGTATGGAGAAGTGGATCCGNGCGGCGCAAGGGGTGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERV3-16A3_I | ZNF460 | 1575 | 1590 | - | 20.91 | CCCTCAGCCTCCCAGA |
| ERV3-16A3_I | ZNF816 | 785 | 799 | + | 20.86 | TGGGGACGTGCTGGG |
| ERV3-16A3_I | scrt | 696 | 706 | + | 18.77 | GCAACAGGTGC |
| ERV3-16A3_I | ANT | 142 | 157 | - | 18.30 | ACACATTTTCCCAGGT |
| ERV3-16A3_I | ZNF331 | 2911 | 2920 | - | 18.17 | TGCAGAGCCC |
| ERV3-16A3_I | ZNF331 | 4596 | 4605 | - | 18.17 | TGCAGAGCCC |
| ERV3-16A3_I | TCP13 | 2185 | 2195 | - | 18.14 | ATGGGGACCAC |
| ERV3-16A3_I | PLAG1 | 1874 | 1887 | + | 18.04 | GGGGGCATATGGGG |
| ERV3-16A3_I | TCP17 | 2185 | 2195 | + | 17.95 | GTGGTCCCCAT |
| ERV3-16A3_I | TGA10 | 3860 | 3869 | - | 17.79 | GCTGACGTCA |
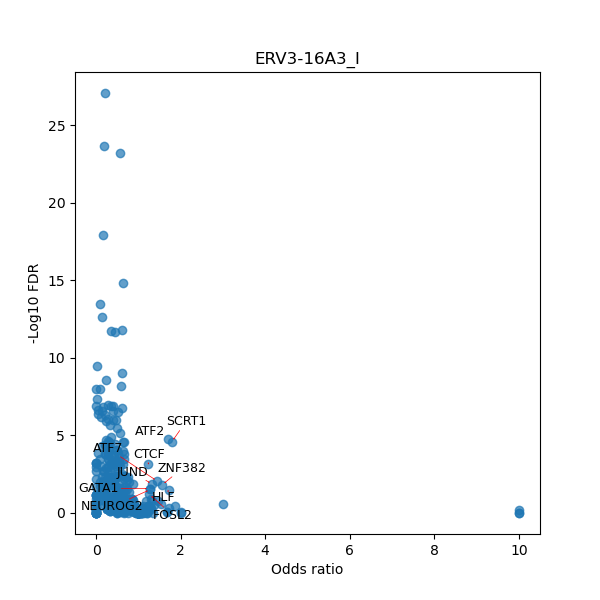
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.