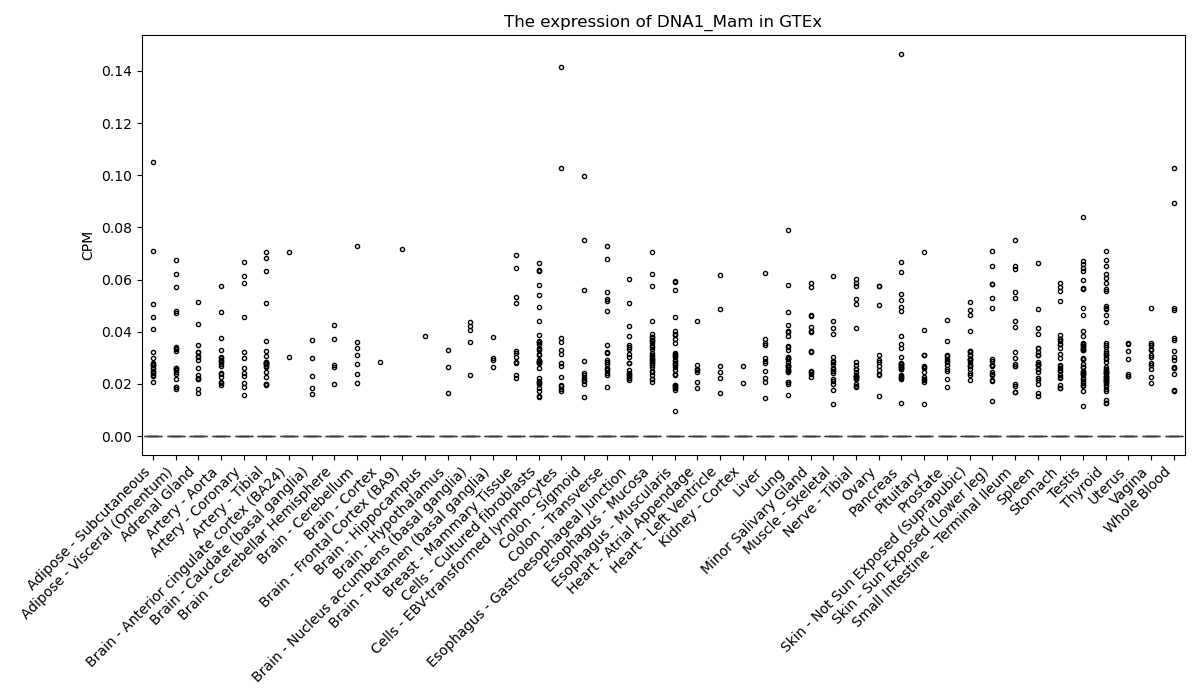
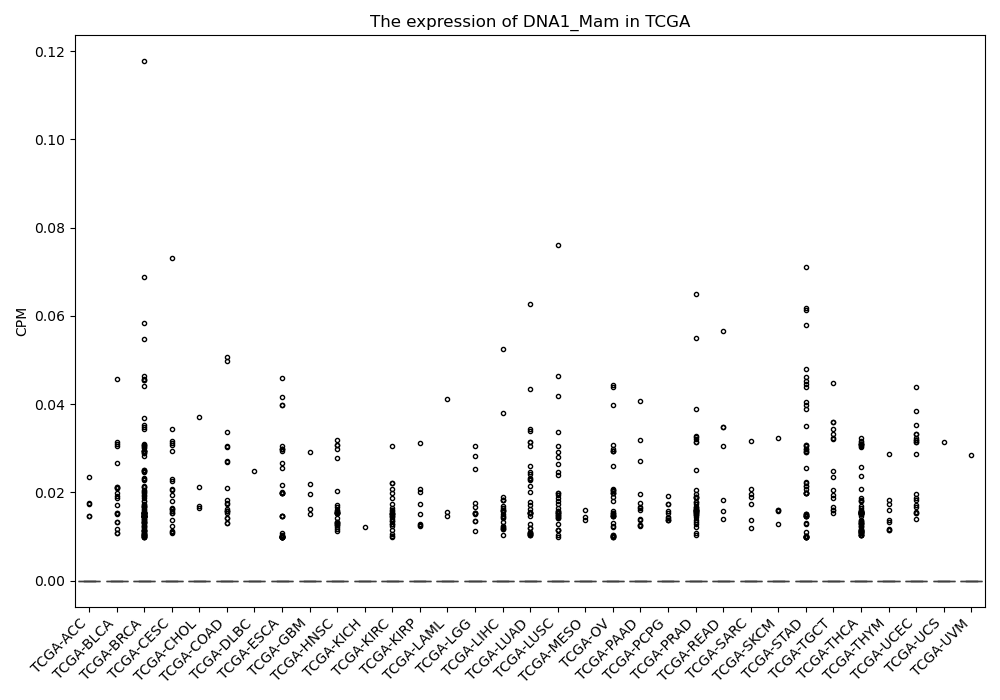
DNA1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000113 |
|---|---|
| TE superfamily | Tc1 |
| TE class | DNA |
| Species | Eutheria |
| Length | 407 |
| Kimura value | 22.80 |
| Tau index | 0.0000 |
| Description | TcMar DNA transposon, DNA1_Mam subfamily (non-autonomous) |
| Comment | Present in ~200 copies in human and other placental mammals. It has imperfect TIRs and putative TSDs (TA, included in consensus). The sequence is a near perfect hairpin. |
| Sequence |
CAGGGTGTCCGAAAAGTCGGGAAACATAGGATAAACTTATTTTTAAACAGTATGTTAGTTACATTTTCAAATAATATGCTCAATATGTTTTTCTTCAACCTCCAGACACCTTTTCAGGTGAAGTACCTCTAAATTTAAAGCAATGGGTCCAATTGTTAATCTGAAAAAAGTACAATAAATACACTATTTTCCCTGTGTTTCCAGACTTTTTGGACACTCTGTAGTGTATTTATTGTACTTTTTTCAGATTAACAATTGGACCCATTGCTTTAAATTTAGAGGTACTTCACCTGAAAAGGTGTCTGGAGGTTGAAGAAAAACATATTGAGCATATTATTTGAAAATGTAACTAACATACTGTTTAAAAATAAGTTTATCCTATGTTTCCCGACTTTTCGGACACCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| DNA1_Mam | SBP8 | 168 | 175 | + | 13.19 | AAGTACAA |
| DNA1_Mam | SBP8 | 233 | 240 | - | 13.19 | AAGTACAA |
| DNA1_Mam | AT2G38090 | 26 | 38 | + | 13.18 | ATAGGATAAACTT |
| DNA1_Mam | AT2G38090 | 370 | 382 | - | 13.18 | ATAGGATAAACTT |
| DNA1_Mam | sma-4 | 102 | 108 | - | 13.18 | TGTCTGG |
| DNA1_Mam | sma-4 | 300 | 306 | + | 13.18 | TGTCTGG |
| DNA1_Mam | Hmga1 | 362 | 369 | - | 12.99 | ATTTTTAA |
| DNA1_Mam | Hmga1 | 39 | 46 | + | 12.99 | ATTTTTAA |
| DNA1_Mam | Hoxd13 | 173 | 179 | + | 12.73 | CAATAAA |
| DNA1_Mam | Hoxd13 | 229 | 235 | - | 12.73 | CAATAAA |
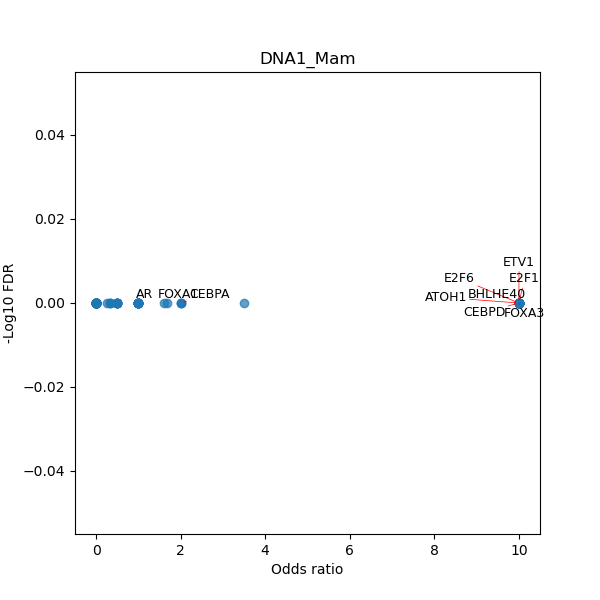
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.