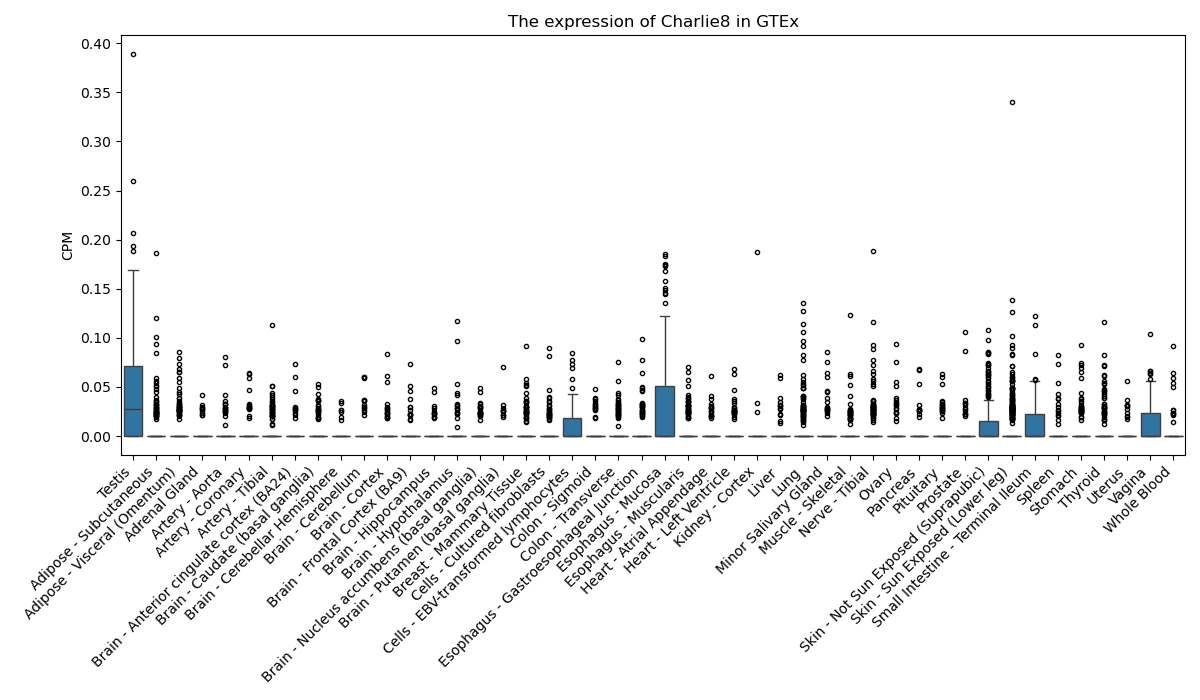
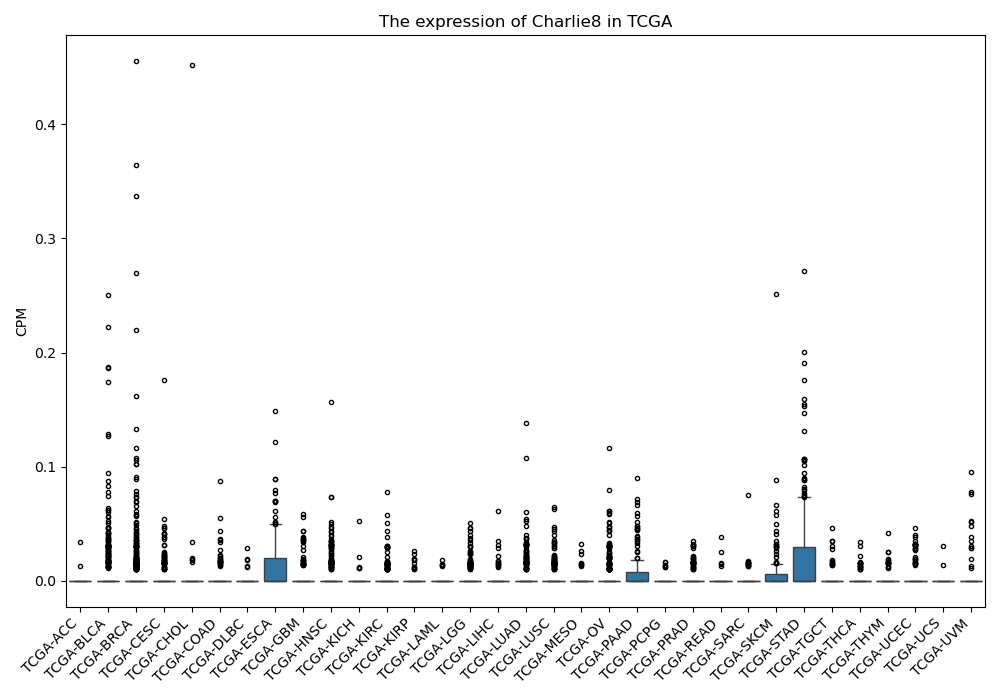
Charlie8
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000107 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2457 |
| Kimura value | 28.91 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie8 subfamily (autonomous) |
| Comment | 15 bp TIR; 8bp TSD with MER1-like NTCTAGAN bias. Nearly complete transposase (roughly <330-2102). |
| Sequence |
CAGAGGTCGCAAACTGGCGGCCCGCGGGCCGNATCCGGCCCGCAGACGTGTTTTGTTTGGCCCGCACAGTGTTTTAAAANATTTTGAATTAGTTGCCAACATTTAAAAATCGGGAGATTTCACATAAAAATCCGGATTTCCGGCTTCTCTTGAAAAATCGGAAGATCTGGCAACACTGGGCCCGCATTCCCGCGTGGCAACAATCGGCTGGAGCTGAGTAGCGGCTGCCCCCTTTAGACGGGGCATGCGCTCTCCAGTTCGCCACAGTCCCCACCACTCCCTATTGTCTCCCCGACACTGAGGCCGAGTGTCAGTTGCCATTTATCATCGCGCTTGCGCTGTTGTTTTTCTTATAGTAGAGTTAAGAGGAAAGTGAAATATTTCTTGTACCCATGTCTCTATCAAAAGTGGGAAAACGAAAGATAGACCGAGAGGGCCGCGTGTTTCAAGAAAAATGGGAGAGAGCATATTTCTTTGTGGAAGTGAAGAATATTCCTACGTGTTTAATATGCAAACAAAGTGTGTCTGTGTCGAAAGAATACAACTTAAGACGCCATTATGAAACAAACCATAGTAAGAACTATGACCGGTATACGGAAAAGATGCGTGATGAAAAACTTAATGAACTGAAAAAAGGACTGAAATTTCAACAGGGTTTGTTTTTGAATGCGAATAAAATAAGTGATGCTGCTGTGAAATGCAGTTATGTATTAAGTGAAAAAATTGCCCGGGCATCAAAACCTTTTACAGATGGCGAGTTTATAAAAGAGTGTCTATTGAATGCAGCAGAAATTATGTGTCCCGAACAGAAACAAGCATTTGCAAATGTAAGTCTAACCGGAAATACTGTTGCTCAGCGTGTTGAAGATACGGCTGAGAACTTACAGGACAAGTTGCGTGAAAAAGTTAAATCATTTGTGGCGTTTTCTATTGCAGCTGATGAGAGCACAGATATAAATAATACCACCCAGTTAGCTATATTTATTCGTGGTGTCGATGAGAATTTTGATGTGACCGAAGAACTTTTGGACATGGTGCCCATGACAGGCACAACATCAGGAAATGACTTATTTTTGTGTGTTGAGAAAAGTCTTGAAAAGTTTAATGTAGACTGGTCAAAATTAGTAAGCGTGACTACAGACGGTGCTCCTGCGATGGTCGGTGTTAACGTCGGACTTGTTACGAAACTTAAATCCAAGGTGGCAACGTTTTGCAAGGACACGGAACTTAAGTCCGTTCATTGCATCATTCACCAGGAATCGCTTTGTGCTAAAAAGTTAAAAATGGAACACGTCATGGACGTAGTAATTAACACCGTGAACTGGATACGCTCCCGTGGCTTGAACCACAGACAGTTCAATGCTTTGCTTGATGAATTGGATGCACAATACGGTAGTCTGCTGTACTACACGGAGGTTAGGTGGCTTAGTCGTGGCGTGGTGCTGAAGAGATTTTTTGAACTGTTGAAAGAAATCGACTTGTTCATGTCATCCAAGGGAAAATCCCCGCCTCGGCTCACCAGCGAAGACTGGATCAAAGACTTGGCCTTTTTGGTTGACATTACAACCCATCTAAACACTTTGAATATTTCTCTGCAAGGGCGTTCACAAGTAGTTACACAAATGTATGATTCGATTCGCTCATTCCTAGCGAAATTGTGTCTTTGGGAAACTCATTTGGCAAGGAATAATCTGGCCCACTTTCCTACGCTGAAATCGGTTTCCAGAAATGAAAGTGATGGCCTGAACTACATTCCCAAAATTGTGGAGTTGAAGACTGAATTCCAAAAAAGGTTCTCTGATTTCAAACTTTATGAAAATGAACTAACATTGTTCAGTTCGCCTTTCTCAATTAATATTGATAGTGTGAATGAAGAGCTACAAATGGAAGTTATTGAACTGCAGTGCAATACGGTACTGAAAACTAAATACGACGACGTTGGAATACCAGAATTCTACAAATATCTCGGGAGTAGTTACCCTAAATATAAAAACCATTGTGCAAAGATTCTATCCATGTTCGGAAGTACCTACGTTTGTGAACAGCTGTTTTCCATTATGAAACTGAATAAAACTAAATATCGCTCCCAGTTAAAGGATTCAAGGCTGAATTCTGTACTGCACATCGCAACGCGAAATACAAGACCTGACATTGACATCTTGGTGCAGAAGAAAAGGTGTCAGATTTCTGGTTCCAAATCAAAGGAGTAACGAATTACGAAAGAAAAATTTAATAATTAAATATTTCTATTTTTCAATTTGTATTTTTTCAATTTCGAATTTAAAATATAATCTCCAATATTATACATGTTTGTATTATGTTACATGCTTACGCCATAGTTCAGTAAAAATAAAGTTTGATTATATTTCTGGCAGTTGATTTTTCTTACACCTGGCCCGCTTCACTCATTTACGTTACCTGCCTGGCCCCTGTAGGCATTTGAGTTTGCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie8 | CAD1 | 1121 | 1131 | - | 16.48 | CGCTTACTAAT |
| Charlie8 | ETV2 | 837 | 845 | + | 16.39 | ACCGGAAAT |
| Charlie8 | YAP7 | 1121 | 1131 | + | 16.26 | ATTAGTAAGCG |
| Charlie8 | DOF5.1 | 1272 | 1290 | + | 16.21 | AAAAAGTTAAAAATGGAAC |
| Charlie8 | PHL11 | 487 | 496 | - | 16.05 | GGAATATTCT |
| Charlie8 | PHL11 | 487 | 496 | + | 16.03 | AGAATATTCC |
| Charlie8 | atfs-1 | 1240 | 1249 | - | 15.96 | ATGATGCAAT |
| Charlie8 | DOF3.4 | 1272 | 1288 | - | 15.89 | TCCATTTTTAACTTTTT |
| Charlie8 | PHL7 | 484 | 496 | + | 15.77 | TGAAGAATATTCC |
| Charlie8 | GT-4 | 566 | 577 | - | 15.65 | TTACTATGGTTT |
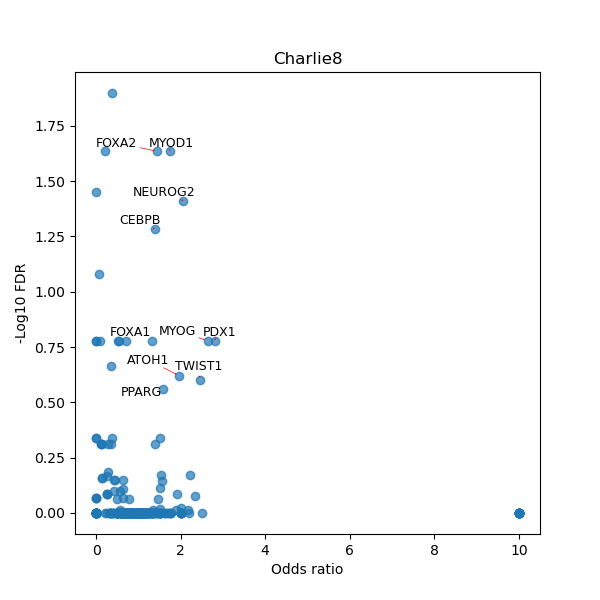
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.