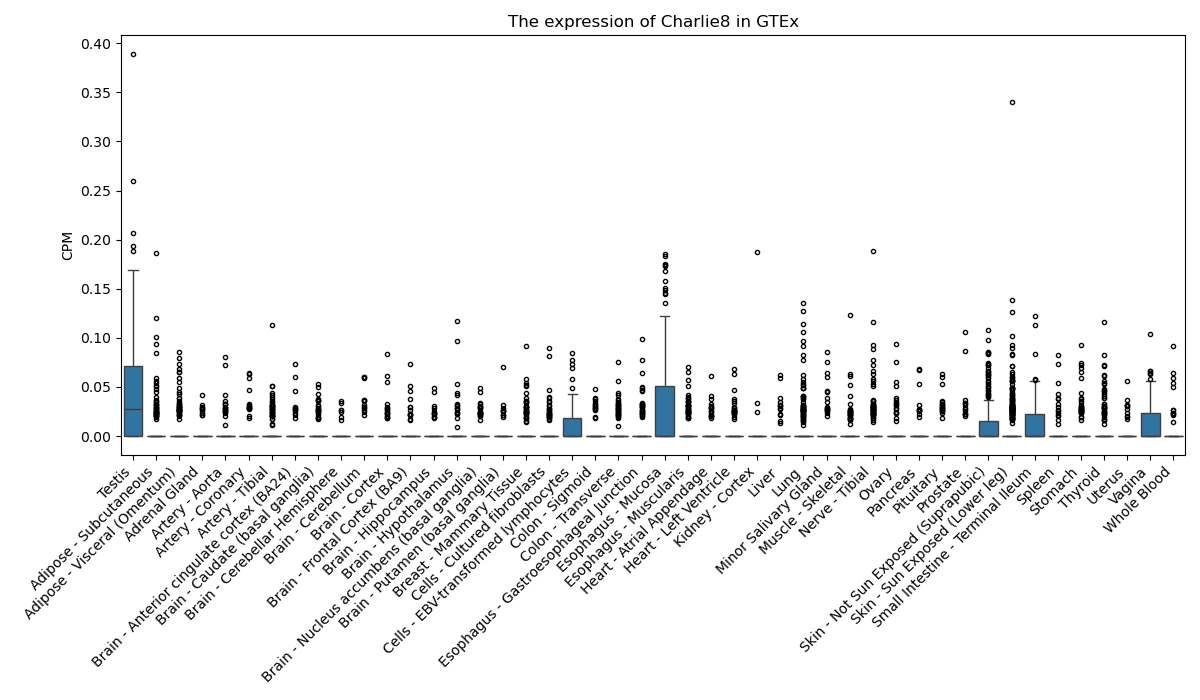
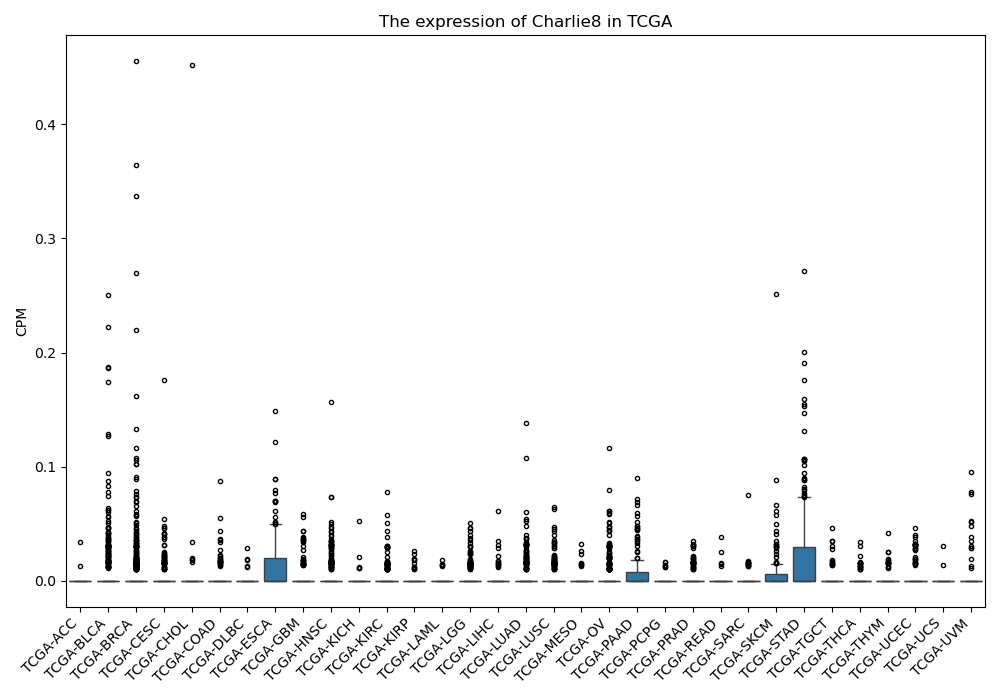
Charlie8
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000107 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2457 |
| Kimura value | 28.91 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie8 subfamily (autonomous) |
| Comment | 15 bp TIR; 8bp TSD with MER1-like NTCTAGAN bias. Nearly complete transposase (roughly <330-2102). |
| Sequence |
CAGAGGTCGCAAACTGGCGGCCCGCGGGCCGNATCCGGCCCGCAGACGTGTTTTGTTTGGCCCGCACAGTGTTTTAAAANATTTTGAATTAGTTGCCAACATTTAAAAATCGGGAGATTTCACATAAAAATCCGGATTTCCGGCTTCTCTTGAAAAATCGGAAGATCTGGCAACACTGGGCCCGCATTCCCGCGTGGCAACAATCGGCTGGAGCTGAGTAGCGGCTGCCCCCTTTAGACGGGGCATGCGCTCTCCAGTTCGCCACAGTCCCCACCACTCCCTATTGTCTCCCCGACACTGAGGCCGAGTGTCAGTTGCCATTTATCATCGCGCTTGCGCTGTTGTTTTTCTTATAGTAGAGTTAAGAGGAAAGTGAAATATTTCTTGTACCCATGTCTCTATCAAAAGTGGGAAAACGAAAGATAGACCGAGAGGGCCGCGTGTTTCAAGAAAAATGGGAGAGAGCATATTTCTTTGTGGAAGTGAAGAATATTCCTACGTGTTTAATATGCAAACAAAGTGTGTCTGTGTCGAAAGAATACAACTTAAGACGCCATTATGAAACAAACCATAGTAAGAACTATGACCGGTATACGGAAAAGATGCGTGATGAAAAACTTAATGAACTGAAAAAAGGACTGAAATTTCAACAGGGTTTGTTTTTGAATGCGAATAAAATAAGTGATGCTGCTGTGAAATGCAGTTATGTATTAAGTGAAAAAATTGCCCGGGCATCAAAACCTTTTACAGATGGCGAGTTTATAAAAGAGTGTCTATTGAATGCAGCAGAAATTATGTGTCCCGAACAGAAACAAGCATTTGCAAATGTAAGTCTAACCGGAAATACTGTTGCTCAGCGTGTTGAAGATACGGCTGAGAACTTACAGGACAAGTTGCGTGAAAAAGTTAAATCATTTGTGGCGTTTTCTATTGCAGCTGATGAGAGCACAGATATAAATAATACCACCCAGTTAGCTATATTTATTCGTGGTGTCGATGAGAATTTTGATGTGACCGAAGAACTTTTGGACATGGTGCCCATGACAGGCACAACATCAGGAAATGACTTATTTTTGTGTGTTGAGAAAAGTCTTGAAAAGTTTAATGTAGACTGGTCAAAATTAGTAAGCGTGACTACAGACGGTGCTCCTGCGATGGTCGGTGTTAACGTCGGACTTGTTACGAAACTTAAATCCAAGGTGGCAACGTTTTGCAAGGACACGGAACTTAAGTCCGTTCATTGCATCATTCACCAGGAATCGCTTTGTGCTAAAAAGTTAAAAATGGAACACGTCATGGACGTAGTAATTAACACCGTGAACTGGATACGCTCCCGTGGCTTGAACCACAGACAGTTCAATGCTTTGCTTGATGAATTGGATGCACAATACGGTAGTCTGCTGTACTACACGGAGGTTAGGTGGCTTAGTCGTGGCGTGGTGCTGAAGAGATTTTTTGAACTGTTGAAAGAAATCGACTTGTTCATGTCATCCAAGGGAAAATCCCCGCCTCGGCTCACCAGCGAAGACTGGATCAAAGACTTGGCCTTTTTGGTTGACATTACAACCCATCTAAACACTTTGAATATTTCTCTGCAAGGGCGTTCACAAGTAGTTACACAAATGTATGATTCGATTCGCTCATTCCTAGCGAAATTGTGTCTTTGGGAAACTCATTTGGCAAGGAATAATCTGGCCCACTTTCCTACGCTGAAATCGGTTTCCAGAAATGAAAGTGATGGCCTGAACTACATTCCCAAAATTGTGGAGTTGAAGACTGAATTCCAAAAAAGGTTCTCTGATTTCAAACTTTATGAAAATGAACTAACATTGTTCAGTTCGCCTTTCTCAATTAATATTGATAGTGTGAATGAAGAGCTACAAATGGAAGTTATTGAACTGCAGTGCAATACGGTACTGAAAACTAAATACGACGACGTTGGAATACCAGAATTCTACAAATATCTCGGGAGTAGTTACCCTAAATATAAAAACCATTGTGCAAAGATTCTATCCATGTTCGGAAGTACCTACGTTTGTGAACAGCTGTTTTCCATTATGAAACTGAATAAAACTAAATATCGCTCCCAGTTAAAGGATTCAAGGCTGAATTCTGTACTGCACATCGCAACGCGAAATACAAGACCTGACATTGACATCTTGGTGCAGAAGAAAAGGTGTCAGATTTCTGGTTCCAAATCAAAGGAGTAACGAATTACGAAAGAAAAATTTAATAATTAAATATTTCTATTTTTCAATTTGTATTTTTTCAATTTCGAATTTAAAATATAATCTCCAATATTATACATGTTTGTATTATGTTACATGCTTACGCCATAGTTCAGTAAAAATAAAGTTTGATTATATTTCTGGCAGTTGATTTTTCTTACACCTGGCCCGCTTCACTCATTTACGTTACCTGCCTGGCCCCTGTAGGCATTTGAGTTTGCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie8 | crp | 2040 | 2051 | - | 17.48 | AAACAGCTGTTC |
| Charlie8 | CEBPG | 1240 | 1249 | - | 17.31 | ATGATGCAAT |
| Charlie8 | sage | 2042 | 2052 | - | 17.25 | AAAACAGCTGT |
| Charlie8 | PHL1 | 485 | 496 | + | 16.88 | GAAGAATATTCC |
| Charlie8 | PHL4 | 487 | 496 | + | 16.80 | AGAATATTCC |
| Charlie8 | DOF5.8 | 1272 | 1290 | - | 16.73 | GTTCCATTTTTAACTTTTT |
| Charlie8 | TFAP4 | 2041 | 2050 | + | 16.65 | AACAGCTGTT |
| Charlie8 | TFAP4 | 2041 | 2050 | - | 16.65 | AACAGCTGTT |
| Charlie8 | TSO1 | 2271 | 2285 | + | 16.65 | AATTTCGAATTTAAA |
| Charlie8 | TCX2 | 2266 | 2280 | + | 16.58 | TTTTCAATTTCGAAT |
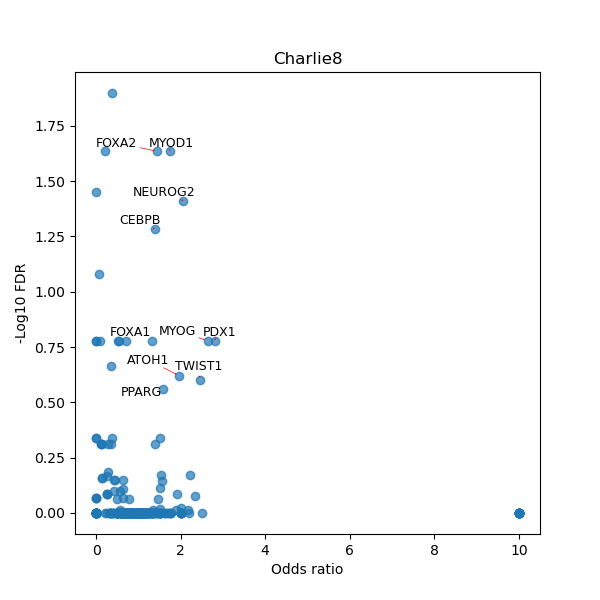
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.