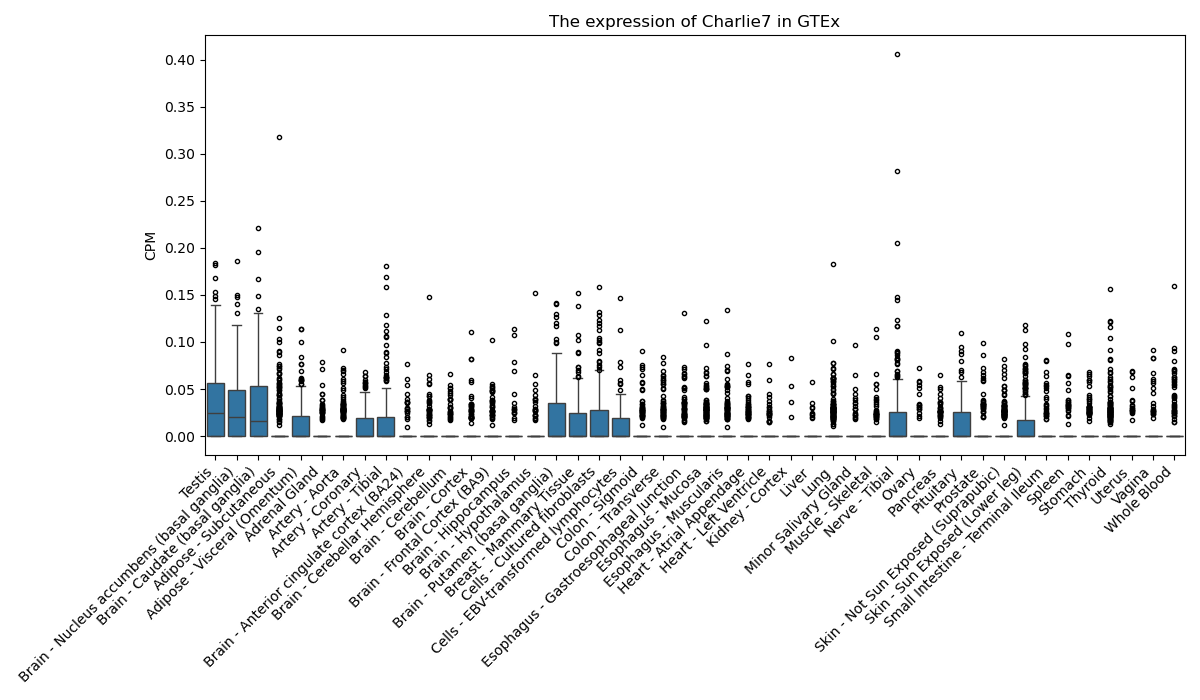
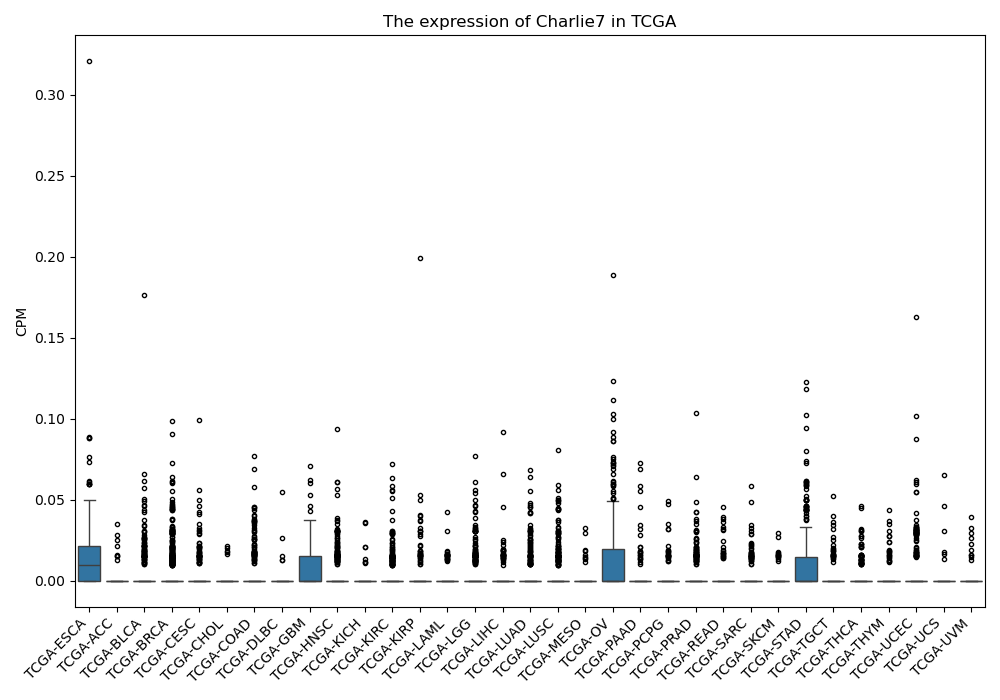
Charlie7
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000627 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Metatheria,Eutheria |
| Length | 2615 |
| Kimura value | 27.16 |
| Tau index | 0.9672 |
| Description | hAT-Charlie DNA transposon, Charlie7 subfamily |
| Comment | 14bp TIR. TSD: NTCTAGAN. ORF at roughly pos. 591-2354. |
| Sequence |
CAGTGGTCTCCAAAGTGGGGTGCGTGCACCCCAGGGAGTGCGCAAGACGATCCATTGGGGTGCGGGAAGAAAATATTAGAACTTCTATTTATATTTATTTTTNATCTAAAAAATAAGAAAGAAATTAAGCTTTACTAATATTTAATATACGGATTGACACTGGCGCCCTCACTCGGTCCGTATGTCAGACGGTCACGTGTCACATGCGGTACGCGAGGTATCCTGAGGGAAGAGTGGGAGTTCCACAACGCGGAGGGGTTGACAGTGGCGCCCTCACTCGTTCGTTCGCTTTCAGCGTATTGCGACGTATTGCAGTTTACGTGTGCCCGGTTAAGTGGATTTACGGATTATATTATCTAGTTTTAACTAAACTAACCCTCACAAAATGGACAAGTGGCTTAAAAAGATTCCTGCAAAGAAACCGCGGATTGAAGATAATACTAATAATGCAAGCACAAGCGAACAACAAGAAAATGGCAGAGCTGACACTTCTACTCCTAGTACGAGCTCTTCGTCAGCCACATTACGAGGTAAAAACAATGACGATCTAATCAGATCTGACAAGAAGTCGGCCAAAAAAATTCGAAATTATCAAGAAGACTATTTGAAATATGGATTTACATCCACTATCGTTAACGATGAACCTCGCCCTAAGTGTATATTGTGCCTTGAGATATTAGCTAATGATAGTATGAAGCCATCACGATTAGCAAGACATTTAAAAACTAAGCATCCAGAACATGAAGACAAACCTCTACAATTTTTTCAGCGATGTTTAAAGTCATGCGATACTCAATCCAGTACTTTACAAAATTTCACTAAACTTAATGATAAATGTTTAGAAGCCTCTTTTGAGGTTTCTTACTTAATAGCGAAAGACAAAAAGCCACATACCATTGGGGAAACACTTGTTCTTCCTGCCGCGGTAAAAATGGCTGAAATAATACACGGAAAACAATATGGCGACAAACTAAAATGCATTCCTTTGTCAGCAAATACTGTTGGAAGACGCATAGAAAACATTGCTGAAGATTTGAAGAAACAAGTATTAGAACAAATTACGCAGTGTGGGAGGTTTGCTATACAGTTGGATGAAAGTACAGATGTTTCTAACATGTCTCAGCTTATGGTATTTGCTAGATTCTGTTTCAATAATGAAATACACGAAGAACTACTTTTTTGTGAGCCACTAAAGGAAAGATGTACCGGAGAAGATATATTCTCAACAGTAAATGACTTCTTTAATAAAAACAATGTTTTATGGGAAAACTGTGTAAGTGTAACCACTGATGGAGCGGCTGCTTTGACTGGAATAAAAAAAGGATTCCGGGGTAAGGTTACAGAGATAGCACCACACGTGAAATTCATTCACTGCATCATTCATAGGCAAGCTATTGCAGCAAAGAAGTTGAAGCCAGAAGTGCACAAAGTGCTACAGGATGTCATCGATGTGGTTAATTTTATAAAAACAAGACCTTTAAAATATAGTAGAATCTTTACAATACTTTGTAATGAGATGGGGAGTGACCATGAAAATCTTTTGTACCACACAGAGGTTCGCTGGTTATCTCGTGGCAAAGTACTTAAAAGAGTTGTCGAACTTAAAGATGAGTTACGCATTTTTCTTTTACAAAAAGACAAGTGTTCCAAATTTGCTGACCTTTTCTGTGATGACAAGTGGCTGTCAGTAGTATGCTACCTAGCAGATATTTTTGAAAAAATAAACACACTTAATCTGTCCCTTCAAGGTAAAGGTGACGTTTTAACAATGAGTGAGAAAGTAACTGCTTTTCGAAAGAAACTCGTGCTATGGAGAGAGCATTTTGAAAACGGATGTTTGGAAATGTTTCCATCGTTATGTGATTTTGTTGCCGAAAACGATGTAAATGTGTCACCTATAAAAACTCTCATATCTGCACACTTTAAAAACTTGGAAACAGAATTTTCTAACCTGTTTAAAAATCTTCCAAATGAAGAGTTTCAGTGGGTTTTGAACCCATTTGTTAAAAATATAAAAATGCAACACCTTCCGATTAGTTTGCAAGAACAACTGATTGACATCAGGGAAGATGGAAATTTACTAGCCGAATTTCAACAAAAACCTTTGCATAATTGGTGGATGGGATTGAAAAATGAGTATCATGATTTAGTAAGCGCAGCCAACGATGCACTTCTTCCATTTGGATCTACGTATCTTTGTGAGGTATCTTTTTCAGCTATGACAGCCATTAAAACCAAGTATCGAAATAAACTGAACTTAGAACCAGACCTTCGAATCGCTGTATCACAAAGTGTTAAACCAAGATTTTCAAAAATAATGAAGCATATTCAATCACATTGCTCTCACTAAAATATTATTAATAATATTTTAGTGAGAGCAAAAATGTTTTGTACCATTAATAAATAAAATAAAAATTATTTTAAAATATTGTTTATTTCATCTTTATCTCATCCTTTTAAAATTTCTATTTTTGTGTATGTTTTATAATGTACATAATATATTAGTACAGTAGTACATGTATATAATTTATAAATAAATATACATATATTGGGGGTGCATGCTCAAAATTTTTTACTGATGGGGATGCGCGATCAAAAAAGTTTGGAGACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie7 | nhr-6 | 1657 | 1664 | - | 15.08 | AAAGGTCA |
| Charlie7 | sage | 1100 | 1110 | - | 15.08 | GAAACATCTGT |
| Charlie7 | ceh-10::ttx-3 | 363 | 376 | - | 15.08 | GTTAGTTTAGTTAA |
| Charlie7 | ZBTB18 | 1098 | 1108 | + | 15.07 | GTACAGATGTT |
| Charlie7 | SOL1 | 2455 | 2469 | - | 15.04 | ATAGAAATTTTAAAA |
| Charlie7 | DOF5.1 | 2429 | 2447 | - | 15.03 | ATAAAGATGAAATAAACAA |
| Charlie7 | FOXD3 | 1714 | 1727 | + | 15.03 | TGAAAAAATAAACA |
| Charlie7 | SVP | 2463 | 2478 | - | 15.02 | TACACAAAAATAGAAA |
| Charlie7 | bigmax | 191 | 199 | + | 15.00 | GGTCACGTG |
| Charlie7 | GT-3a | 194 | 207 | + | 14.97 | CACGTGTCACATGC |
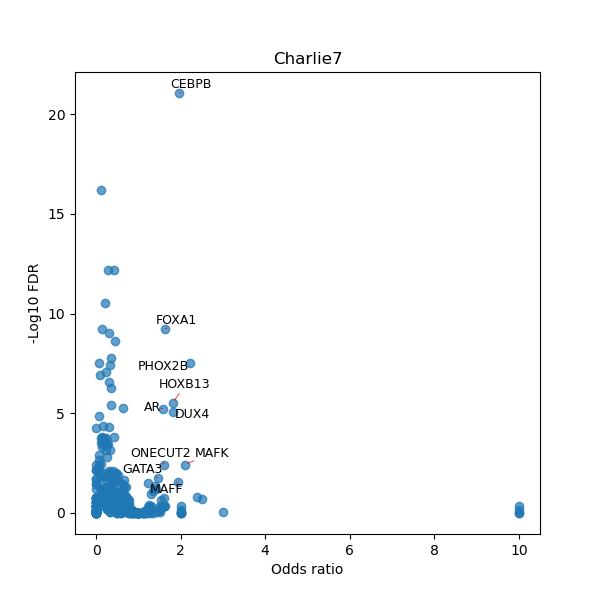
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.