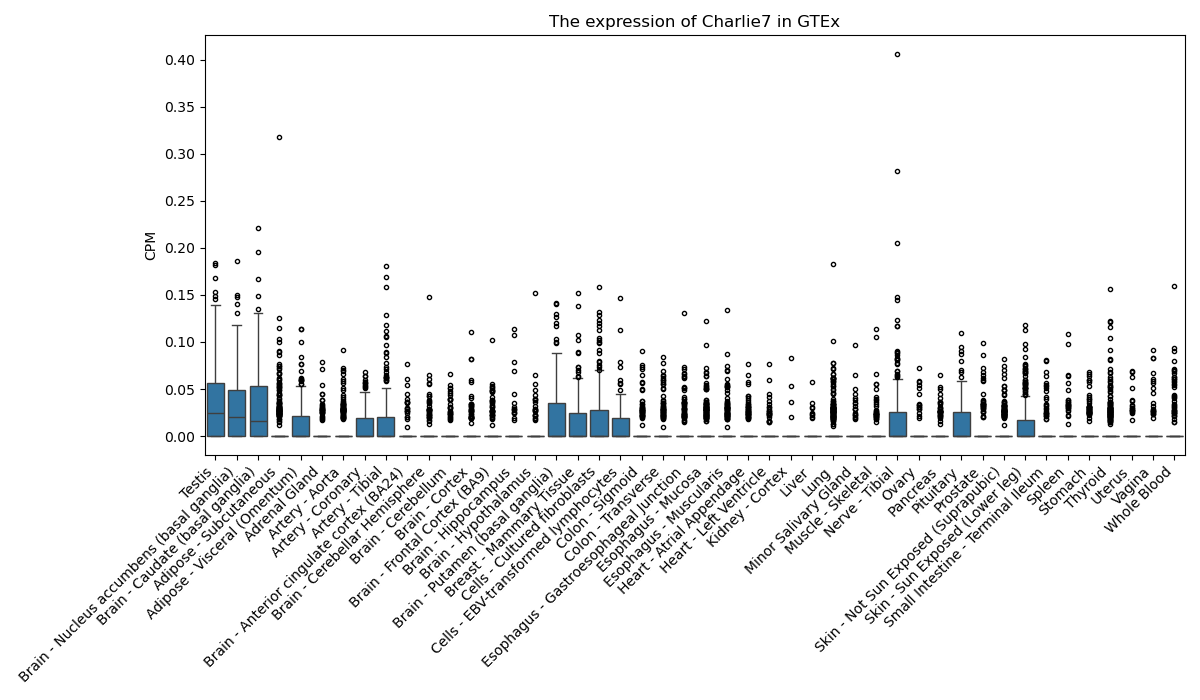
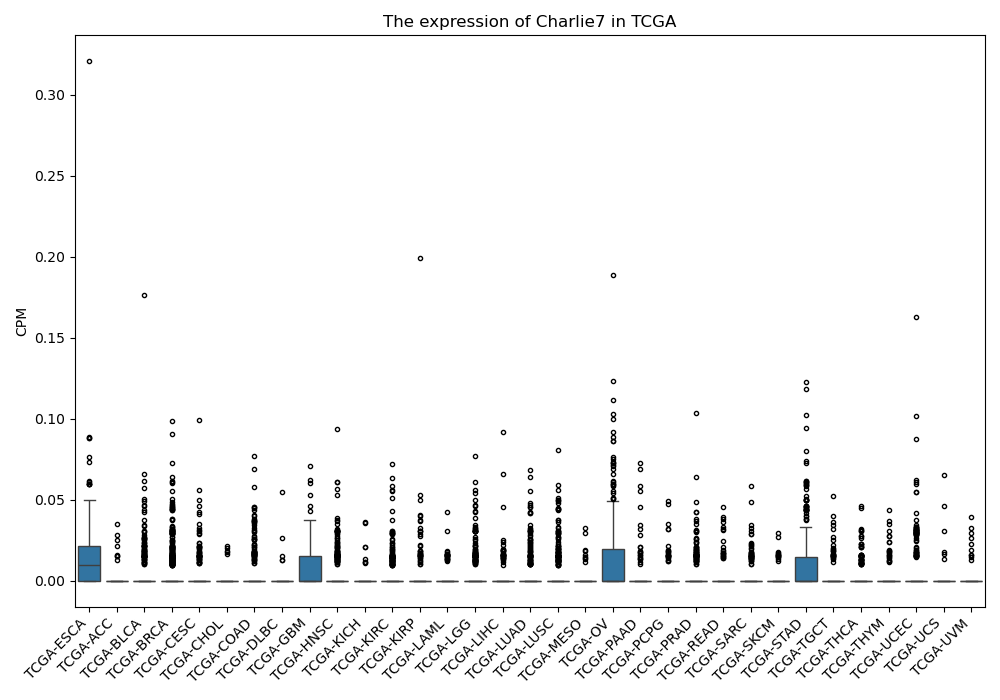
Charlie7
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000627 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Metatheria,Eutheria |
| Length | 2615 |
| Kimura value | 27.16 |
| Tau index | 0.9672 |
| Description | hAT-Charlie DNA transposon, Charlie7 subfamily |
| Comment | 14bp TIR. TSD: NTCTAGAN. ORF at roughly pos. 591-2354. |
| Sequence |
CAGTGGTCTCCAAAGTGGGGTGCGTGCACCCCAGGGAGTGCGCAAGACGATCCATTGGGGTGCGGGAAGAAAATATTAGAACTTCTATTTATATTTATTTTTNATCTAAAAAATAAGAAAGAAATTAAGCTTTACTAATATTTAATATACGGATTGACACTGGCGCCCTCACTCGGTCCGTATGTCAGACGGTCACGTGTCACATGCGGTACGCGAGGTATCCTGAGGGAAGAGTGGGAGTTCCACAACGCGGAGGGGTTGACAGTGGCGCCCTCACTCGTTCGTTCGCTTTCAGCGTATTGCGACGTATTGCAGTTTACGTGTGCCCGGTTAAGTGGATTTACGGATTATATTATCTAGTTTTAACTAAACTAACCCTCACAAAATGGACAAGTGGCTTAAAAAGATTCCTGCAAAGAAACCGCGGATTGAAGATAATACTAATAATGCAAGCACAAGCGAACAACAAGAAAATGGCAGAGCTGACACTTCTACTCCTAGTACGAGCTCTTCGTCAGCCACATTACGAGGTAAAAACAATGACGATCTAATCAGATCTGACAAGAAGTCGGCCAAAAAAATTCGAAATTATCAAGAAGACTATTTGAAATATGGATTTACATCCACTATCGTTAACGATGAACCTCGCCCTAAGTGTATATTGTGCCTTGAGATATTAGCTAATGATAGTATGAAGCCATCACGATTAGCAAGACATTTAAAAACTAAGCATCCAGAACATGAAGACAAACCTCTACAATTTTTTCAGCGATGTTTAAAGTCATGCGATACTCAATCCAGTACTTTACAAAATTTCACTAAACTTAATGATAAATGTTTAGAAGCCTCTTTTGAGGTTTCTTACTTAATAGCGAAAGACAAAAAGCCACATACCATTGGGGAAACACTTGTTCTTCCTGCCGCGGTAAAAATGGCTGAAATAATACACGGAAAACAATATGGCGACAAACTAAAATGCATTCCTTTGTCAGCAAATACTGTTGGAAGACGCATAGAAAACATTGCTGAAGATTTGAAGAAACAAGTATTAGAACAAATTACGCAGTGTGGGAGGTTTGCTATACAGTTGGATGAAAGTACAGATGTTTCTAACATGTCTCAGCTTATGGTATTTGCTAGATTCTGTTTCAATAATGAAATACACGAAGAACTACTTTTTTGTGAGCCACTAAAGGAAAGATGTACCGGAGAAGATATATTCTCAACAGTAAATGACTTCTTTAATAAAAACAATGTTTTATGGGAAAACTGTGTAAGTGTAACCACTGATGGAGCGGCTGCTTTGACTGGAATAAAAAAAGGATTCCGGGGTAAGGTTACAGAGATAGCACCACACGTGAAATTCATTCACTGCATCATTCATAGGCAAGCTATTGCAGCAAAGAAGTTGAAGCCAGAAGTGCACAAAGTGCTACAGGATGTCATCGATGTGGTTAATTTTATAAAAACAAGACCTTTAAAATATAGTAGAATCTTTACAATACTTTGTAATGAGATGGGGAGTGACCATGAAAATCTTTTGTACCACACAGAGGTTCGCTGGTTATCTCGTGGCAAAGTACTTAAAAGAGTTGTCGAACTTAAAGATGAGTTACGCATTTTTCTTTTACAAAAAGACAAGTGTTCCAAATTTGCTGACCTTTTCTGTGATGACAAGTGGCTGTCAGTAGTATGCTACCTAGCAGATATTTTTGAAAAAATAAACACACTTAATCTGTCCCTTCAAGGTAAAGGTGACGTTTTAACAATGAGTGAGAAAGTAACTGCTTTTCGAAAGAAACTCGTGCTATGGAGAGAGCATTTTGAAAACGGATGTTTGGAAATGTTTCCATCGTTATGTGATTTTGTTGCCGAAAACGATGTAAATGTGTCACCTATAAAAACTCTCATATCTGCACACTTTAAAAACTTGGAAACAGAATTTTCTAACCTGTTTAAAAATCTTCCAAATGAAGAGTTTCAGTGGGTTTTGAACCCATTTGTTAAAAATATAAAAATGCAACACCTTCCGATTAGTTTGCAAGAACAACTGATTGACATCAGGGAAGATGGAAATTTACTAGCCGAATTTCAACAAAAACCTTTGCATAATTGGTGGATGGGATTGAAAAATGAGTATCATGATTTAGTAAGCGCAGCCAACGATGCACTTCTTCCATTTGGATCTACGTATCTTTGTGAGGTATCTTTTTCAGCTATGACAGCCATTAAAACCAAGTATCGAAATAAACTGAACTTAGAACCAGACCTTCGAATCGCTGTATCACAAAGTGTTAAACCAAGATTTTCAAAAATAATGAAGCATATTCAATCACATTGCTCTCACTAAAATATTATTAATAATATTTTAGTGAGAGCAAAAATGTTTTGTACCATTAATAAATAAAATAAAAATTATTTTAAAATATTGTTTATTTCATCTTTATCTCATCCTTTTAAAATTTCTATTTTTGTGTATGTTTTATAATGTACATAATATATTAGTACAGTAGTACATGTATATAATTTATAAATAAATATACATATATTGGGGGTGCATGCTCAAAATTTTTTACTGATGGGGATGCGCGATCAAAAAAGTTTGGAGACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie7 | ZAP1 | 1470 | 1484 | - | -4.62 | ATTTTAAAGGTCTTG |
| Charlie7 | THI2 | 2453 | 2467 | - | -4.66 | AGAAATTTTAAAAGG |
| Charlie7 | NR1H2::RXRA | 2251 | 2267 | - | -5.24 | CTGGTTCTAAGTTCAGT |
| Charlie7 | RARA | 181 | 198 | + | -5.26 | TATGTCAGACGGTCACGT |
| Charlie7 | TRP1 | 2469 | 2489 | - | -5.91 | ATTATAAAACATACACAAAAA |
| Charlie7 | GLIS1 | 11 | 25 | - | -9.36 | ACGCACCCCACTTTG |
| Charlie7 | AT4G12670 | 337 | 365 | - | -16.55 | TAAAACTAGATAATATAATCCGTAAATCC |
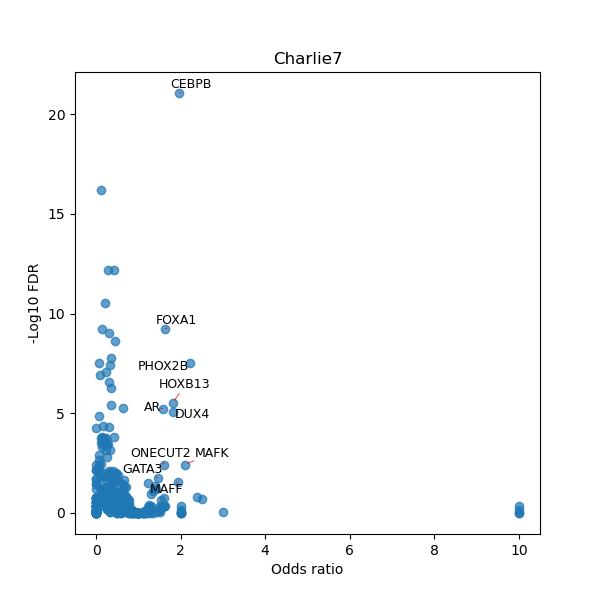
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.