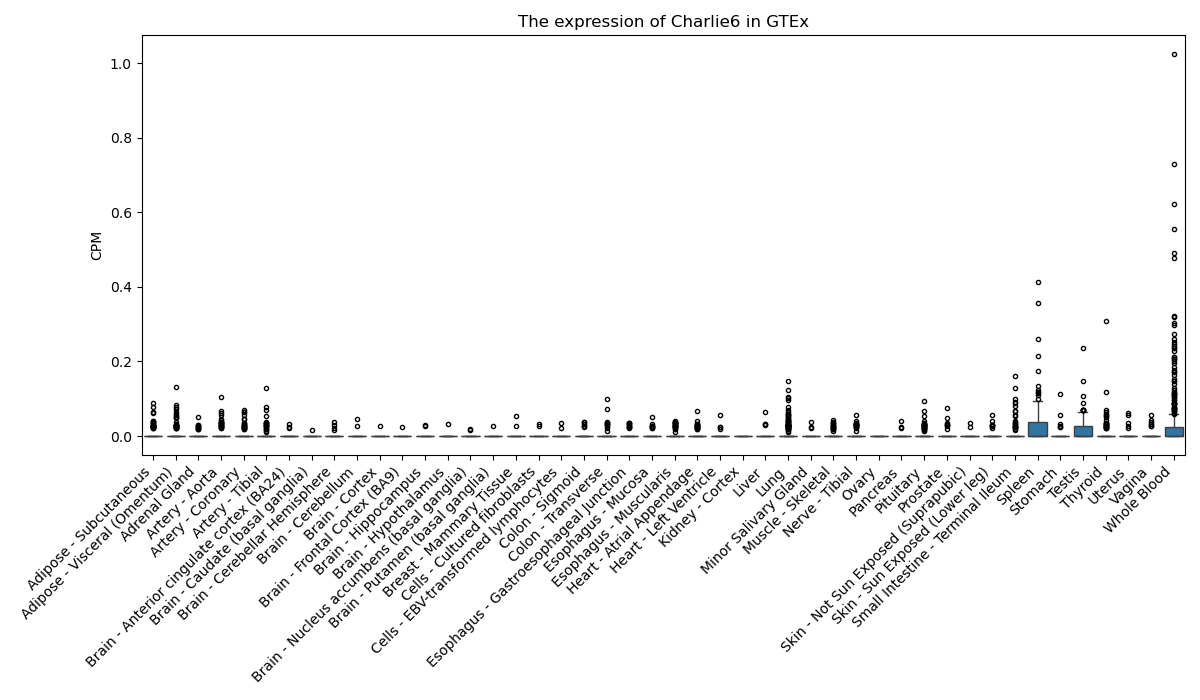
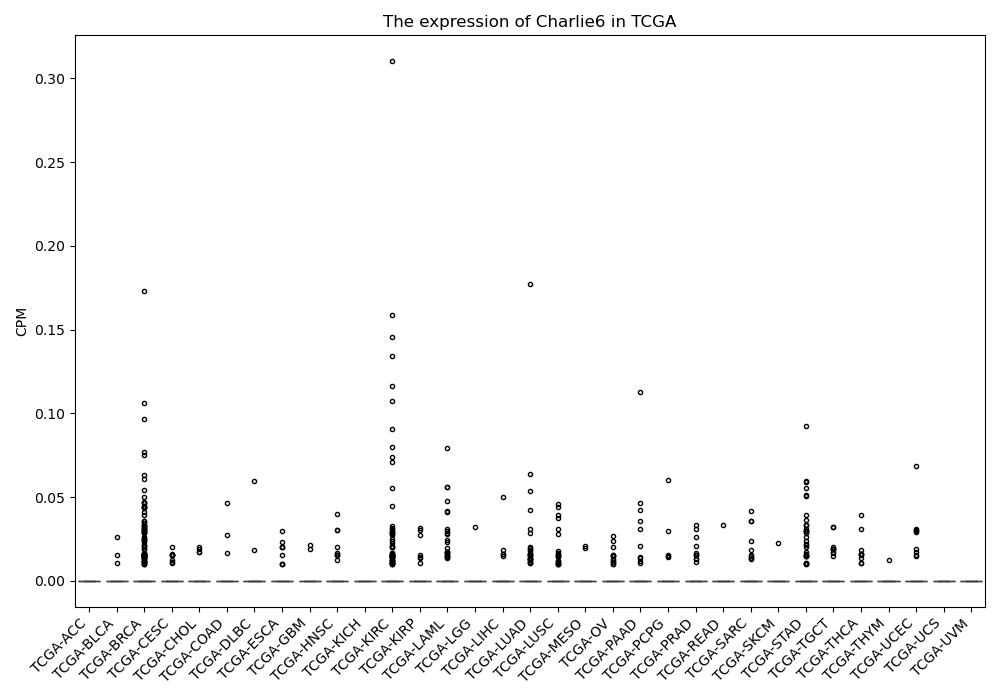
Charlie6
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000105 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 3519 |
| Kimura value | 23.01 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Charlie6 subfamily (autonomous) |
| Comment | Imperfect 16bp TIR. TSD: NTCTAGAN. Charlie6 is unusual in that no common deletion products have been amplified. A member of the MER1-group. ORF roughly at pos. 607-2496. |
| Sequence |
CAGAGTTTCTCAAATGGGGTTTCTGCGGAACCTAGGGTTCCGCGAGATGTCGCTAGGGGCTCCGCGAGANATTGTGATTGAAAAAAATATAGATTTTTGAACTTCGCGCGCCGCGCGATGCTAGCACTCGCAGTGATGGACAGTGACCGAGCAGCCCCGTTAGCAATTTCGTTAGAGGTCTTTCAGCCTCGTATGGCAGTGCGCAGTAGGACCATGTTCATTTGGNTGAGTCCATTCTCAGTTTTTGACGCAAGATAGGGGAGGCCGTTGATAANTGGTGAGTGCTGTATTGCACGGAGGGGAGGACGGTAGGCTGATGACGTCAGCCTCTCCCTCCCGAATTACCTCCCCCAACCTCCCCTTNCGCGCCGCCGACTGACTTACGGGAGCGAACAAGCTCTACCGGTGTAATAGAAAATCGGAGGGAAATTTTCATTCTAAAGAAGGAATTTTTACGCGCCGCGACTCAGCTGCCCCCCTGCCGCTTTGCTGTTGTTGTAAACGTTGCAAAACGTGGATTGTGTAGGTTAGAAGTGAATTGTATTGTGAAGTTTTCGTATTTTTTGCGATTTTCTCGTTTAGTTATTTATTATGCCCTTCTGTTTTATATTTTATTAGCGTTTGTGTTTTACAAACATGTCTGACGGTCCAATGTGGCGATTTTTGAAGACGAGGTGTGAGACGGAAGAGGAGGATTCTAGGCCTAGGCCTACGGACAATTCTGATAAGGTTAGTGATGAAGAATTACAATCTTCTGAGTCATTTCACTGCACTAGTGCAACAAGGGACACAAAAAAAGTCCGTCTTTACAATGAAAGCTACTTATCAATGGGTTTTACGTGGNCTGGTAATCCAAGTTGTCCTATTCCATTGTGCGTCGTCTGTGGCAAACAACTTACAAATGCAGCAATGGCTCCAGCAAAATTGAAAAGACACTTAACTACAAATCACAGCCATTTGACAAGTAAAGGTGCTGATTATTTTAAACGGCTATTGGAATCTCAAAACAAACAGAGTAAAGCTTTTGTTNAAAAAGTCACATTCAGTGAAAAGGCTCAGGAAGCAAGTTATTTAGTAGCAGAACTTATTGCCCAGAAAAGGAAAAGTCACACAGTTGGTGAGAACCTAATAATGCCAGCATGTAAAATTATAGTGAGTAAAATGCTAGGACAAGATGCAGTACGAGAAATTGAAAAGGTTCCACTCTCAAACAGTACGATAAGTCGACGTATTGATGACATGTCACATGATGCTGAAGAGGTTTTGTGTAATAAACTGAAAAACAACAGCTTCTCTATCCAGGTTGATGAGTCAACAGATTTCACCAATAAATGTCATGCTGTAGCATTTGTAAGATTTGTAAATGATGGTGAAATTCAAGAAAACTTTTTCTGCTGCAAAGAGCTGCCCGAAACAAGCAAAGGCCAAGATATATTTAATGTTTTGTCTTCATATCTGGAAACAAAAGGTCTGTCTTGGAGGAACTGTGTTGGCATCTGTACTGATGGTGCCCCATCAATGGTTGGCTCCATGAGAGGTTTCGCCTCTCTTGTAAAAAAAGAAAATCCTGACGTTGTCACAACACACTGCTTTCTTCACAGAGAGGTGCTGGTGTCAAAAACTCTTGGAGATGAAATGAAAAAAGTTCTGGATGATGCTACAAAAATGGTTAACTTTATTAAACAAAGACCAGTTCACTCGAGAATGTTTNAAAAACTGTGTGAAAACCTGGACAAAGAGCACATAAATCTCCTGCTACATACAGAAATCCGGTGGCTTAGCAGAGGAAGAGTTCTCAACAGGGTGTTTGAGCTGAAAGGTGAATTGCAGGAGTACTTTCAAGAAAATAGTAGGCCAGATTTTGCTGAGTGCTTTGAAGATGAAGAATGGCTGCAGAAACTAGCCTACTTAGCAGACATTTTTCATCACATGAACCAGTTGAACAAGTCTCTGCAAGGCCCTGGAGAAAATGTTTTGACTTCAAGTGACAAGATTCTTGGATTTAAAAGGAAACTGAATCTTTGGAAAAATCATGTTGCAAAAGGAAATCTTGAAATGTTTCCACTGCTGCTTGGGCTTGAGAGTGAGGAAGGATATCAGCAAGTCTCAAGTCTTATTGAAAACCACCTGGAAGAACTGCAGAACAAAATTGAACAGTATTTTCCCTCCCTTTCAACACAAGTGTATGACTGGGTGAGGGACCCTTTCTCTGAATCTTCTGCTCAGCCTGAGAACTTGACTTTGAGAGAAGAGGAAGAACTTTGTGAGCTGCAGTCTGATCGTACACTCAAGATGAGATTTACTGATCTGCCCCTAGACGAGTTCTGGATTTCTGTGAAAGAAGAGTATCCTGCCATTCACAGGAAAGCAATGAACATTTTGCTGCAGTTTTCAACTTCTTACATGTGTGAGCAAGCTTTTTCTTGTTTAACAAGCATCAAGAGCAAGGACAGAAATCGTCTCATTTCAGTTGAAAATGAAATCCGTGTGTGCTTATCTCAAGTTCGACCCAGAATTGAGTATTTGTGCAGCAAAAAACAAGCACAGGTTTCACATTAAAGAGGTAAATTTCATTCTTTATTTTTAGTTTCAAAAATAACACTTTTTATGCATATATAGGCCTATAAAAGAGATCAAGAAACCAACACATGTGTATAATAATTTTGTAACTGCTAGGCCTATATTTGAGCCTGATCATGTCACTCAGTAAGAAAATATTTTTTCAAAGTCTTATATAAAATTGTGTCTTAGGCTATATTTTTATTCATATTGCTTTGGCTTATGGTTTTAGTAGCTTTACTATTCAACATTGTCAATAAATTTTCATGTTGTAAATAACATTAATTAAAATCAATTTACAACCTTTATTTAAATTAAATCTACCGAGCCTACCTTTAGAAAAATTAAATCCCATTTTGGCTTAAGCCAGTTATTTAACAGAAATTTATTATGTTTGCCTTATGAGTTTTTAAAATTTATGAAAAGGAAAGAAATTAATTTCAAGAAAGGTAAGCTAAGCTTAATACCTTNTTTTTTTCAAGAAAGNTTGGCTAAAAAGTCATTTTTAGTTTTAAATAGGGAGAAAAACTACTCTTGCACATACCATATACCACATACCATACCTCATTAGCATCTTGTATAGTAGAAATTTGTGATAAGACTTTTAGGCCTACTTGGATAAGGTNAAAAAATCAAGTAGGAAATTTTTTAAAAAAGAAAAACTGAAGTTTCCTCTACCTACACTACTGACTTGCATTAGAGTTGATATCCTTTGTGGGGAGGGGGAACCACACAAAAAGAGTAGGCTAATAAGTTGGCCAAAATAAACAGATCCACAGAAAAATATCATTCTCTACTCAAAAGAGCATTGACAATTATTATTGGATTATTATATCTACAACTTACTGATCACGCTAATGTACCATGAGCTGTAGATATAATAATTTTTATGCAGGGGTTCCCTGAGACCTGAAAATTATTTCAAGGGCTCCTCCAGGGTAAAAAGATTGAGAAAGGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie6 | Znf423 | 27 | 41 | + | 19.70 | GGAACCTAGGGTTCC |
| Charlie6 | NAC020 | 2420 | 2440 | - | 19.41 | TGATGCTTGTTAAACAAGAAA |
| Charlie6 | NAC075 | 2420 | 2439 | - | 19.15 | GATGCTTGTTAAACAAGAAA |
| Charlie6 | Prdm15 | 1723 | 1733 | + | 18.89 | GAAAACCTGGA |
| Charlie6 | TGA3 | 317 | 327 | + | 18.88 | ATGACGTCAGC |
| Charlie6 | TGA9 | 317 | 327 | - | 18.68 | GCTGACGTCAT |
| Charlie6 | NAC058 | 2420 | 2438 | + | 18.55 | TTTCTTGTTTAACAAGCAT |
| Charlie6 | TGA4 | 315 | 325 | - | 18.45 | TGACGTCATCA |
| Charlie6 | NAC013 | 2420 | 2438 | + | 18.43 | TTTCTTGTTTAACAAGCAT |
| Charlie6 | NAC005 | 2420 | 2438 | + | 18.33 | TTTCTTGTTTAACAAGCAT |
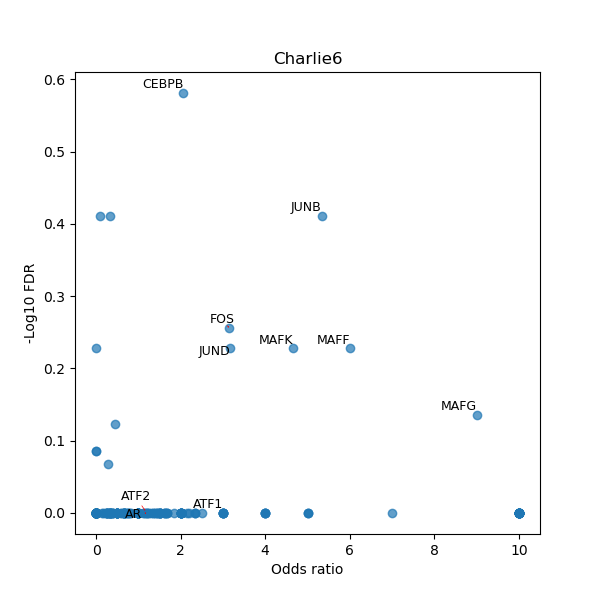
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.