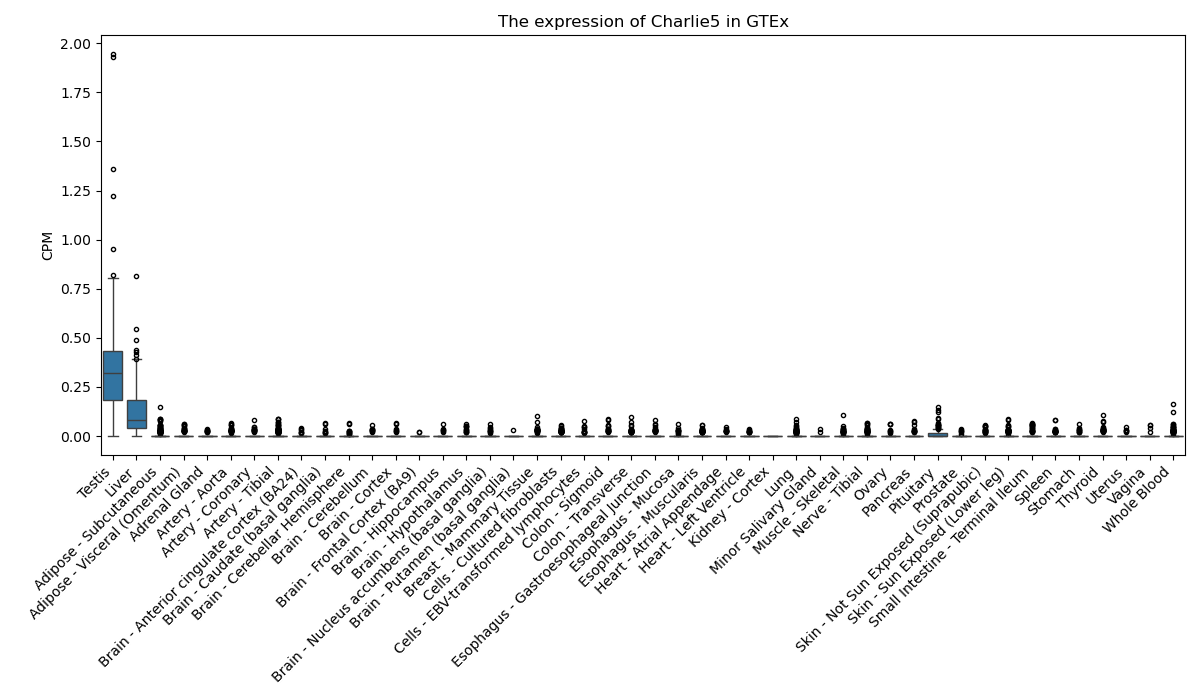
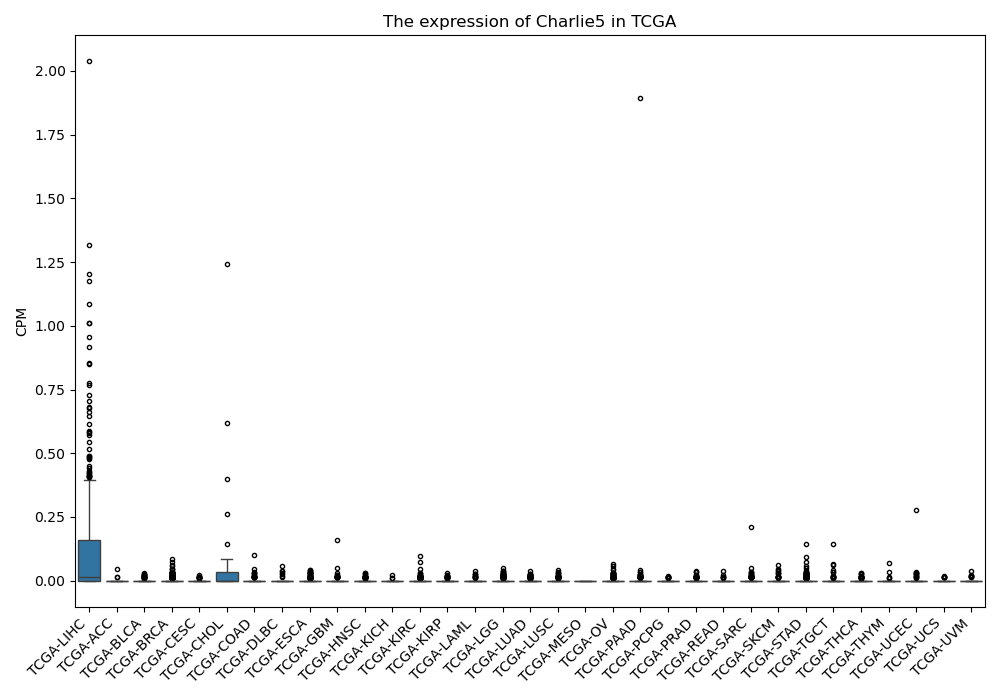
Charlie5
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000027 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2624 |
| Kimura value | 17.76 |
| Tau index | 0.9941 |
| Description | hAT-Charlie DNA transposon, Charlie5 subfamily |
| Comment | Transposose (roughly <841 to 2439) is 50% similar to the human protein AC004883. MER3 appears to be a recombinant product of 5' end fragments of Charlie5. |
| Sequence |
CTGCGCTGTCCAATACGGTAGCCACTAGCCACATGTGGCTATTGAGCACTTGAAATGTGGCTAGTGCGACTGAGGAACTGAATTTTTAATTTTATTTAATTTTAATTAATTTAAATTTAAAAACTGAAGCAGTGTAAAATATTTTTCCGTTAAACACAACTTTATTGTTTTGGTAGGACTACATTTCACTTTAACCGTTGCATCGCGTAAGATATTATTGTTGTATTGTAGTGCGCGTGTCGGGCACGTGCGTCGTTTCTAGTATTACACATAAACACATCACCGATCTAGTCGGTGTCAACGGATTGATTCAGTTCGAATGATTTTTTTCTACGCACCGATGTAACATTGTAATGTGTTTATTTGAATATTTTATGCAGACAGCACGAGTTACAGTGATACCTATGTAAATCGTAATTGGTAATTAAATTGAAATACTTATTATTTTAATTATAATATAATTAAATTATTTTTCTAGTTTAAAAAATAAGTATGGACAAATTTTTAAATTTAAAACGAAGGCATACTACTGATGCAGAATCGAGTGGAGATGCAGAAGCTAGTACTACGACCGGAACAGTAAAGAAAAAAAGAGACTGGAAGAAGGTATGTCGCAAATTTCACGATGAATGGCAATTGCAATTTGCTGCGGCAGAGCAAAACGAAAAAGCTGTTTGTTTGTTGTGTAAAAAAATTTTTAAAGATAATAAAGTGGACAATATTAAGAGACATTTTCAGCAAATACATAGTGAATTTGATAAGAAGTTTCCTCTCAACAGTCAAAAAAGAATCGATGAAATTAGTCGCCTGAAATCAGAATTAAATGTCCAACAAAAATTTTTTAAATAATTTTTAACAGGATCTGAGCTTGTAACTTTGGCCAGCTATAAAACGGCTTGGATTCTTGCACAAAAAAGAAAACCATTTTTAGATGGAGAGATGGTAAAANAAATTATTATTTCAGTTATGGAAATTTTGTTAGAAAATTATGAGGAAAAGACTAAAAAAGATATTTTACAAAAAGTGAAAGATCTTCAATTAAGCCACCAAACAATTGCCCGTAGAATACAAGACCTTTCTAACAATATCAAAGATCAATTGATTCAAAATTTGAAAAATTGCAAGTACTTTTCTTTAGCTTTAGATGAGTCGTGCGATATAAGAGACACTGCCCAATTAATACTTTGGGTACGTTTTGTCTCAAAGGACTTCCAAATTTACAAAGAAATGTTGNCAATTCGCGGCCTAAAAAATCGAACTCGTGGCGTAGATATTTTTNAATCTTTTACATCTGTCAAAGAAGAATTTCAGCTAGATATGAAAAAATTAGTTTCTATCACGACGGACGGTGCTCCAGCTATGTTAGGTCAAAAATCCGGATTTATTGGAATTTTAAAACAAGAGACTGATGTTTCCCTTATTGCTTCGTTCCACTGTATGATACATATTGAAAATATTTGTGCTCAGTTTTCTGAAGCAGACTCTATGAAAAGTGTCATGGATACAGTTGTTAAAATCGTTCAGTATATACGTGCAAATGCTATGAATCATCGCCAGTTTATGGAACTGTTGAAAGAAATAGAAGACAATGAATTTAATGATCTTGTGTTCTTTGCCAATGCTCGTTGGTTGAGTCGTGGAAGAGTTTTACAAAGATTTACTGTACTGTTAACTCCAATTCAAGATTTTCTTGAAACAAAAGGAATGCTTGCCAAATATTCAATAATCAAAGACAAAAAATGGCAGTGTGATTTACGTTTTCTCACCGATATCACACTGCATATGAACGAGCTAAATTTGAAGCTCCAAGGAAAGGAAAAGCTTATTTGTGACCTAGCTAGACAGGTACGAGAATTTATGTTGAAATTGAAACTTTTCATAATACAAATCAATAATAATGATTTTACACATTTTTCTAACATGAATCAATATGCAGAAGATTTTAATTGTAATCGACAGCATTATGTAAATTGGCTGCAAAAACTACAAGAAAAATTTGAAGAACGCTTTGTTGATATTGATAAATTTAGAGTTGCTTTTCAATTTATGCAATACCCCTTTGAATTCGATGTTAATAATACTGAGTTGACACAAGAGTTAGTGAATTTACTTAACTTGGACAGACGTAGTTTTGAAACTGATATGCTTTTGCTTCAAAGTCAAATCAATTCTTCTAAAAAAGATGAACCAGTTTTGTCAATGTGGATGCGAATATTAAAGGAAAATGATTTTTTGATACTCGATTCAGTTATTGGAAAACTTTTAAGTATGTTTGGAACAACTTGGGTATGTGAATCTACTTTTTCAACTGTAAATTTTATGAAATCTAAATACAGATCAAGTATTTCCGATGAAAATTTAGCGTCCGAATTGAGATGTGCTGTAAGTGTAAAATACACACCGGATTTCGAAGACTTAGTACGAAAAAAAGAATGTAAAATATCTCATTAATAATTTTTATATTGATTACATGTTGAAATGATAATATTTTGGATATATTGGGTTAAATAAAATATATTATTAAAATTAATTTCACCTGTTTCTTTTTACTTTTTTNAATGTGGCTACTAGAAAATTTAAAATTACATATGTGGCTCGCATTATATTTCTATTGGACAGCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie5 | TSO1 | 110 | 124 | - | 17.74 | GTTTTTAAATTTAAA |
| Charlie5 | TCX6 | 362 | 376 | + | 17.69 | ATTTGAATATTTTAT |
| Charlie5 | PROP1 | 459 | 469 | - | 17.61 | TAATTTAATTA |
| Charlie5 | SOL1 | 1102 | 1116 | + | 17.48 | ATTCAAAATTTGAAA |
| Charlie5 | TSO1 | 107 | 121 | + | 17.33 | TAATTTAAATTTAAA |
| Charlie5 | ZNF680 | 1296 | 1306 | + | 17.27 | CAAAGAAGAAT |
| Charlie5 | PHOX2B | 459 | 470 | + | 17.23 | TAATTAAATTAT |
| Charlie5 | PROP1 | 459 | 469 | + | 17.12 | TAATTAAATTA |
| Charlie5 | TCX6 | 502 | 516 | - | 16.85 | TTTTAAATTTAAAAA |
| Charlie5 | DOF3.4 | 2539 | 2555 | + | 16.64 | GTTTCTTTTTACTTTTT |
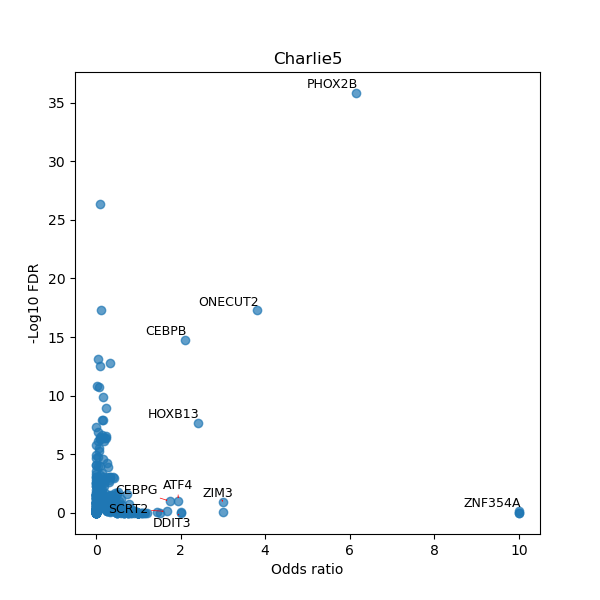
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.