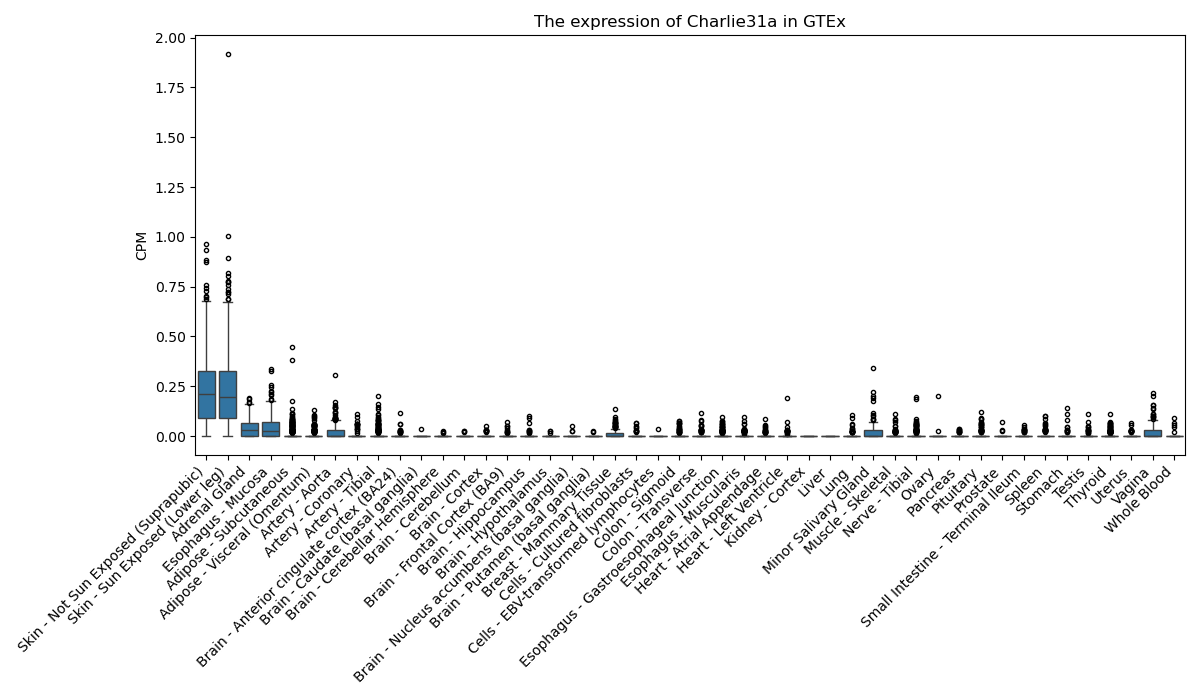
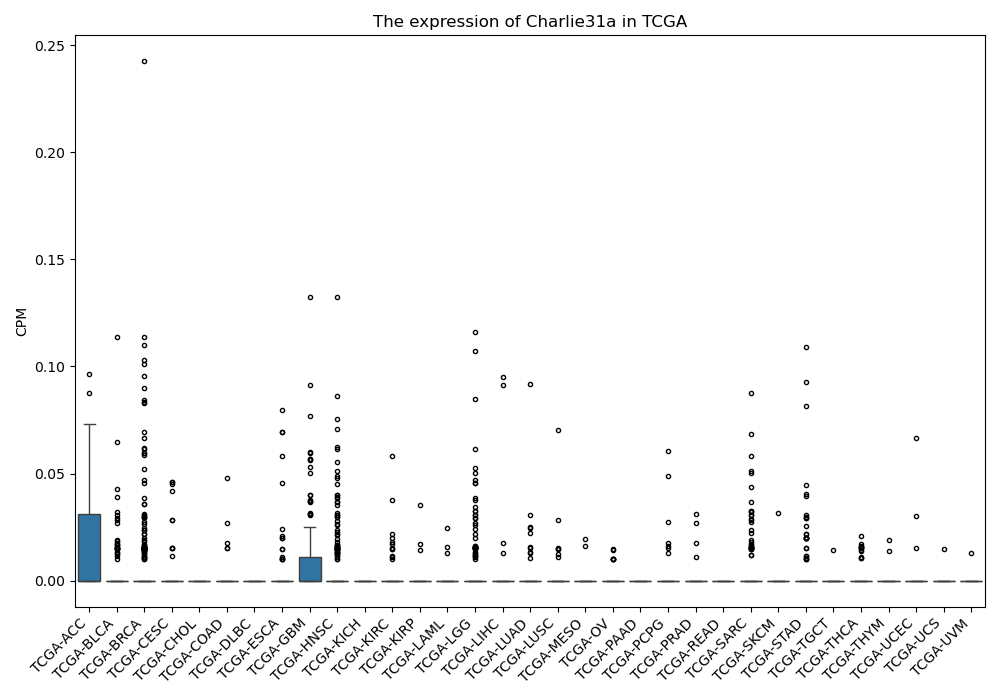
Charlie31a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001244 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 554 |
| Kimura value | 31.04 |
| Tau index | 0.9730 |
| Description | hAT-Charlie DNA transposon, Charlie31a subfamily |
| Comment | 8 bp TSDs with NNCTAGNN preference. Terminal 80-90 bp similar to those of autonomous Charlies (like Chaplin6_FR and Charlie17), on which the orientation is based. Present in elephant, sloth, human, absent in marsupials. |
| Sequence |
CAGTGGTTCTCAACCGGTGTGCCGCGGCACACCGGTGTACCGCGAACCCTTCACAGGTGTGCCGCCAAATTTTGATCAATGTGTAAAATTTATTTTTGCTACAACTGAGTATACATACTGTTACGGCGATGTCTGTCAGACGGAACGTTGAGCTGAGCGGGTCGTGCGACGCGGTGTAGAGCGGGGTACAACTTGCTGCGAGTAAGAGGTAAGAGGCATAGCGTAACGTCAAGCCGGCGCCAAAGAAAGATCGCGTGTTTACTGGAACATGCGAGAACTTTGCCTCGTGAGTATATAAGAGAGCGCGGTCAGGGCAGTAAACAGTTCTACTCAAACAGCGACACAGTGCTGAAGGGCGACCAAGTGAATAGTGCGATAACTGTGGATGCTTTTATAGACACTGTGAGACATTGTTATACTGTAGTAGTGATTGCGTTGTAATTATTGCTGACTTGTTTAGTGTTGTTTTGAGTCTTTTAATCCCTTCAAGTGTGCCGCCAAAAATTCTAGTATTCGAAAGTGTGCCGCGAACACAAAAAGGTTGAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie31a | Os05g0497200 | 235 | 242 | + | 14.24 | CGGCGCCA |
| Charlie31a | HSF4 | 265 | 277 | - | 14.17 | TTCTCGCATGTTC |
| Charlie31a | snai-1 | 53 | 59 | - | 14.14 | CACCTGT |
| Charlie31a | tj | 443 | 452 | - | 14.13 | GTCAGCAATA |
| Charlie31a | ATHB-20 | 440 | 447 | - | 14.11 | CAATAATT |
| Charlie31a | ATHB-53 | 439 | 447 | - | 14.03 | CAATAATTA |
| Charlie31a | Hsf1-2-4 | 265 | 282 | - | 13.98 | CAAAGTTCTCGCATGTTC |
| Charlie31a | ZNF257 | 206 | 215 | + | 13.92 | GAGGTAAGAG |
| Charlie31a | FOXP2 | 255 | 263 | - | 13.68 | AGTAAACAC |
| Charlie31a | NRL | 442 | 453 | + | 13.66 | TTATTGCTGACT |
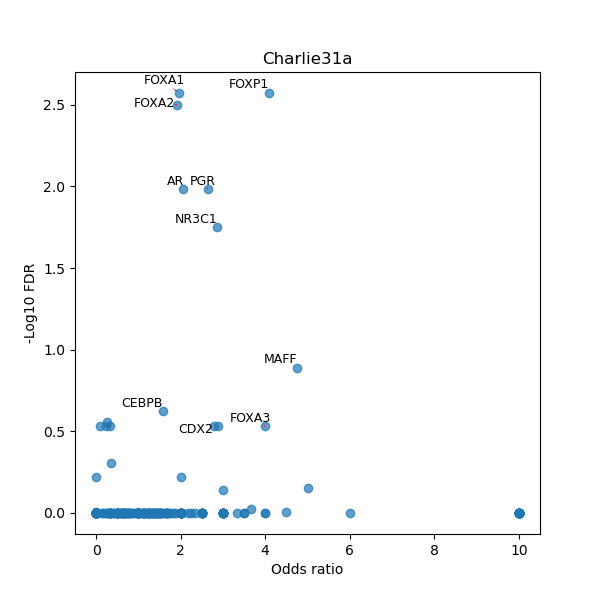
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.