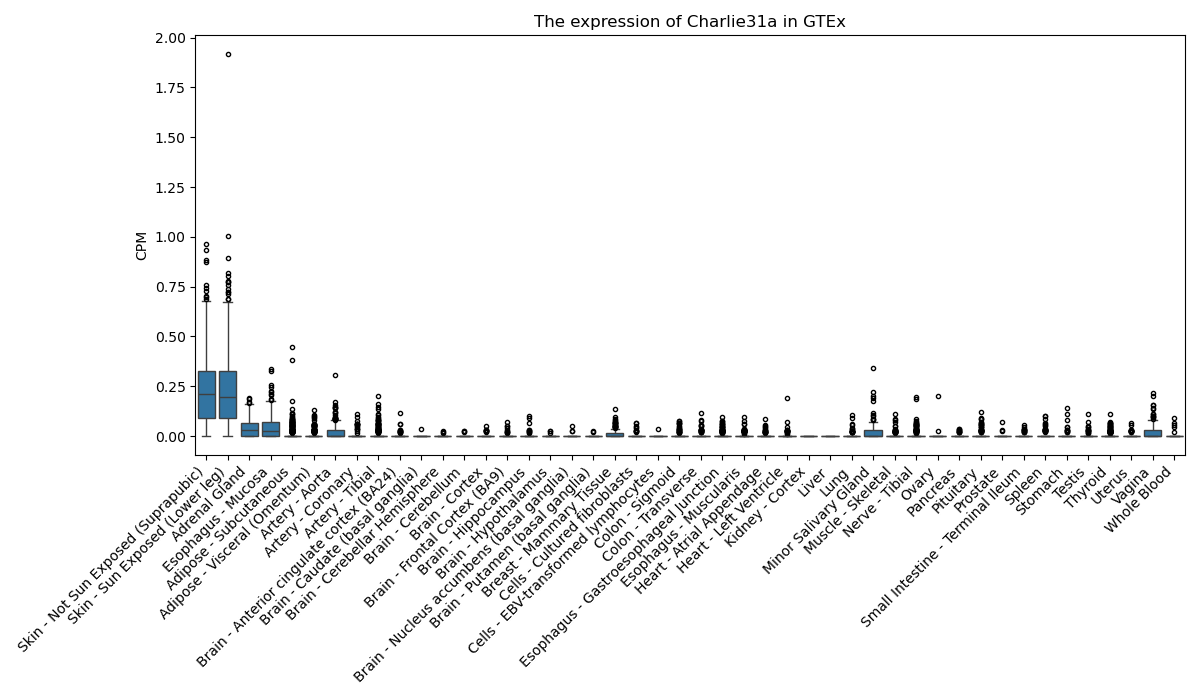
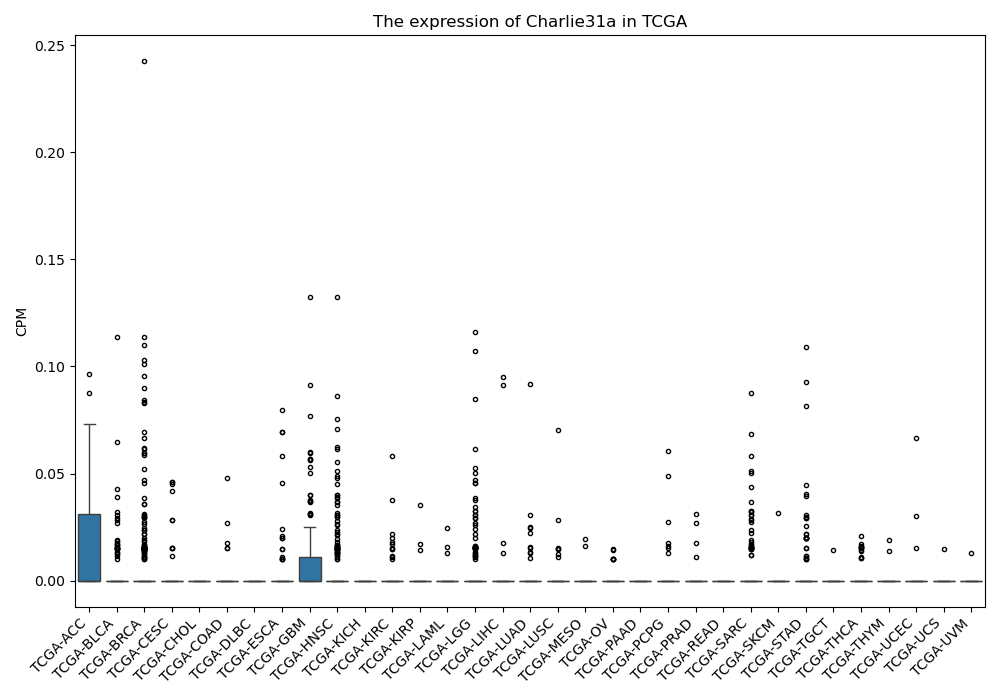
Charlie31a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001244 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 554 |
| Kimura value | 31.04 |
| Tau index | 0.9730 |
| Description | hAT-Charlie DNA transposon, Charlie31a subfamily |
| Comment | 8 bp TSDs with NNCTAGNN preference. Terminal 80-90 bp similar to those of autonomous Charlies (like Chaplin6_FR and Charlie17), on which the orientation is based. Present in elephant, sloth, human, absent in marsupials. |
| Sequence |
CAGTGGTTCTCAACCGGTGTGCCGCGGCACACCGGTGTACCGCGAACCCTTCACAGGTGTGCCGCCAAATTTTGATCAATGTGTAAAATTTATTTTTGCTACAACTGAGTATACATACTGTTACGGCGATGTCTGTCAGACGGAACGTTGAGCTGAGCGGGTCGTGCGACGCGGTGTAGAGCGGGGTACAACTTGCTGCGAGTAAGAGGTAAGAGGCATAGCGTAACGTCAAGCCGGCGCCAAAGAAAGATCGCGTGTTTACTGGAACATGCGAGAACTTTGCCTCGTGAGTATATAAGAGAGCGCGGTCAGGGCAGTAAACAGTTCTACTCAAACAGCGACACAGTGCTGAAGGGCGACCAAGTGAATAGTGCGATAACTGTGGATGCTTTTATAGACACTGTGAGACATTGTTATACTGTAGTAGTGATTGCGTTGTAATTATTGCTGACTTGTTTAGTGTTGTTTTGAGTCTTTTAATCCCTTCAAGTGTGCCGCCAAAAATTCTAGTATTCGAAAGTGTGCCGCGAACACAAAAAGGTTGAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie31a | E2FC | 489 | 503 | + | 16.41 | AGTGTGCCGCCAAAA |
| Charlie31a | lsy-2 | 417 | 424 | - | 15.13 | CTACAGTA |
| Charlie31a | FOXP2 | 316 | 324 | + | 14.61 | AGTAAACAG |
| Charlie31a | bHLH80 | 189 | 197 | - | 14.56 | AGCAAGTTG |
| Charlie31a | ZNF417 | 236 | 242 | + | 14.55 | GGCGCCA |
| Charlie31a | IG1 | 492 | 500 | + | 14.40 | GTGCCGCCA |
| Charlie31a | IG1 | 59 | 67 | + | 14.40 | GTGCCGCCA |
| Charlie31a | Ptf1A | 53 | 60 | - | 14.30 | ACACCTGT |
| Charlie31a | bHLH130 | 189 | 196 | + | 14.26 | CAACTTGC |
| Charlie31a | daf-16 | 255 | 262 | - | 14.26 | GTAAACAC |
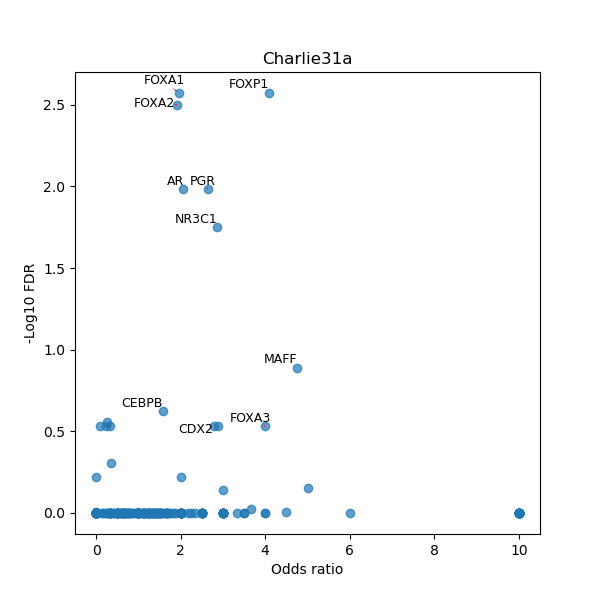
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.