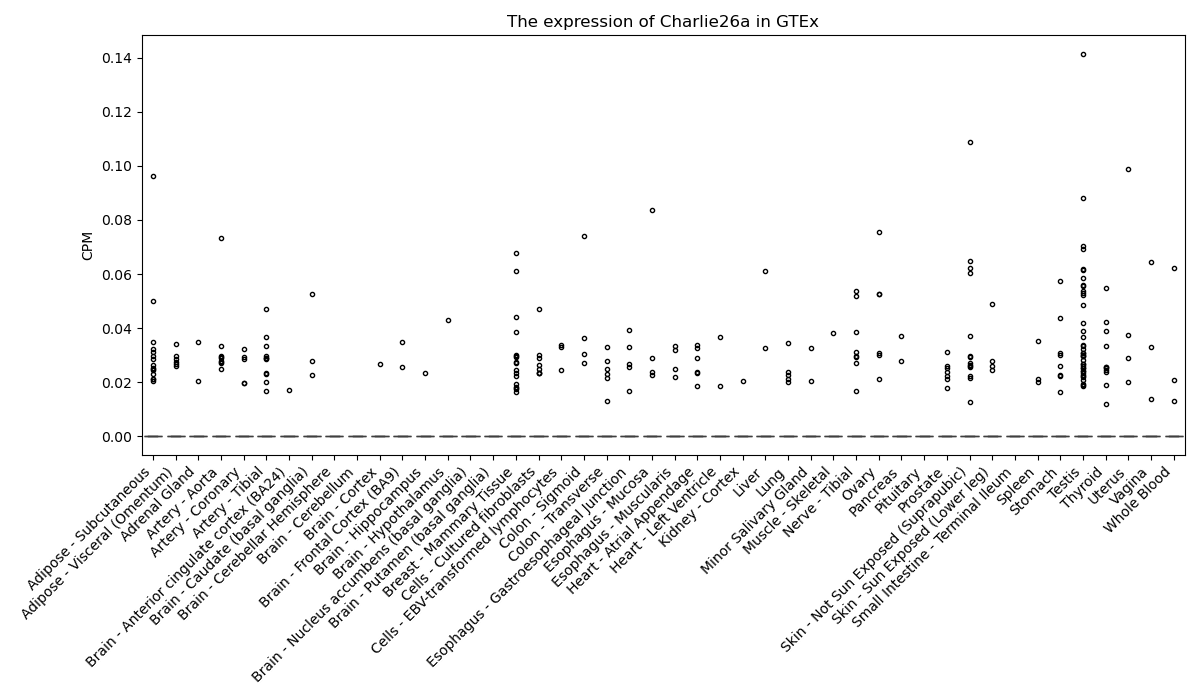
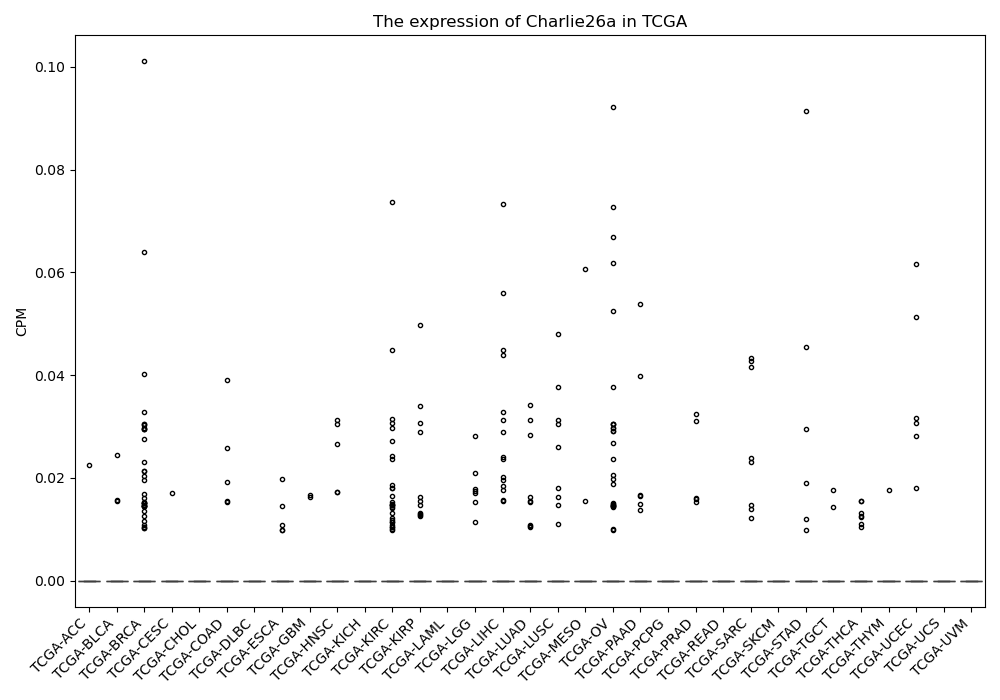
Charlie26a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000103 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 325 |
| Kimura value | 28.01 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Charlie26a subfamily (non-autonomous) |
| Comment | 14 bp TIR. |
| Sequence |
CAGTGATGAGCAACCCGCGGCCCGCCCGGCTTCGCGATACGGCCCGCGATCTAATTTCAGGATGAAAGATTAGAAAGCTGCCTCCGTGTTGCTACTTCTCAAATATGCCCAGATATTAACATTTTAGTGGCTAAAAAGCAGTGCCAATTTTCTCATTGACATTCTTCTATTCAATAAAGTAGGTAATCTAAGTTGTAAGAATATACATTTTCCCCCATGGGACTCAATAAAGCTATTTTCATTTTGAATGAAAAAAAAATGCGGCCCGTAAACATTTTTATTTTTCCTGAATTGGCCCTCGTGCAAAAAAAGTTGCTCACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie26a | ZNF416 | 106 | 115 | - | 17.29 | TATCTGGGCA |
| Charlie26a | BAD1 | 15 | 26 | + | 15.11 | CCGCGGCCCGCC |
| Charlie26a | ETV2::HOXB13 | 277 | 289 | - | 14.96 | CAGGAAAAATAAA |
| Charlie26a | KLF15 | 20 | 27 | + | 14.65 | GCCCGCCC |
| Charlie26a | MCM1 | 284 | 295 | - | 14.29 | CCAATTCAGGAA |
| Charlie26a | PK02532.1 | 111 | 118 | - | 14.19 | TAATATCT |
| Charlie26a | FOXA1 | 268 | 275 | + | 14.06 | GTAAACAT |
| Charlie26a | F10B5.3 | 282 | 292 | + | 14.00 | TTTTCCTGAAT |
| Charlie26a | ZBED4 | 21 | 30 | + | 13.82 | CCCGCCCGGC |
| Charlie26a | Ebf2 | 214 | 222 | + | 13.62 | CCCATGGGA |
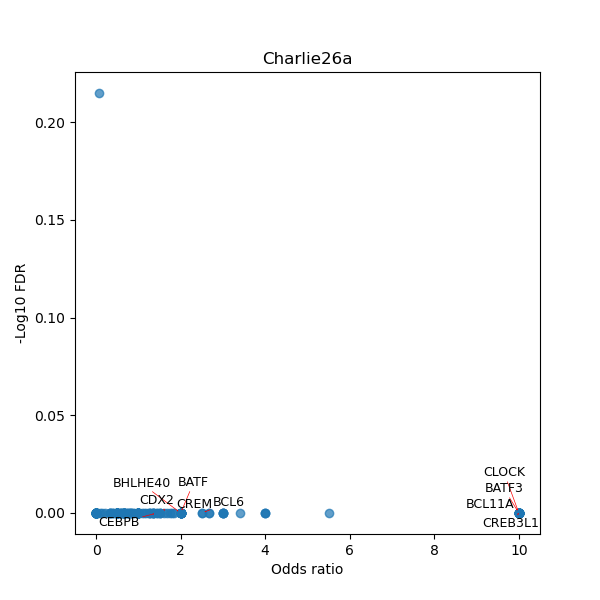
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.