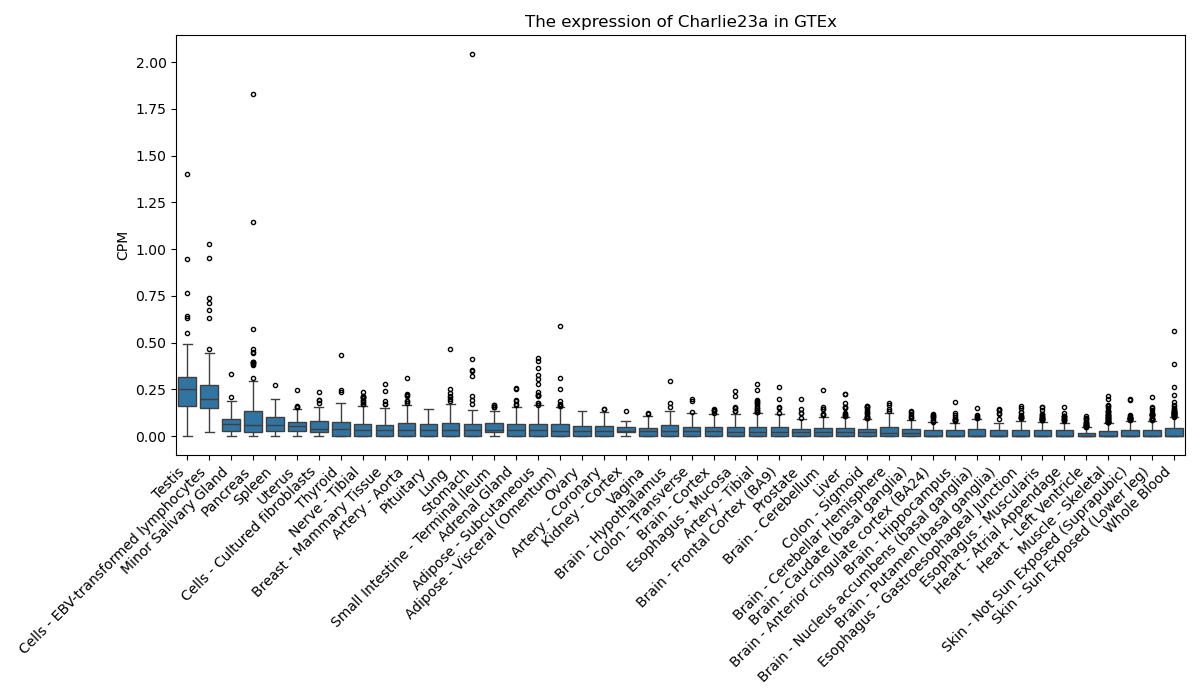
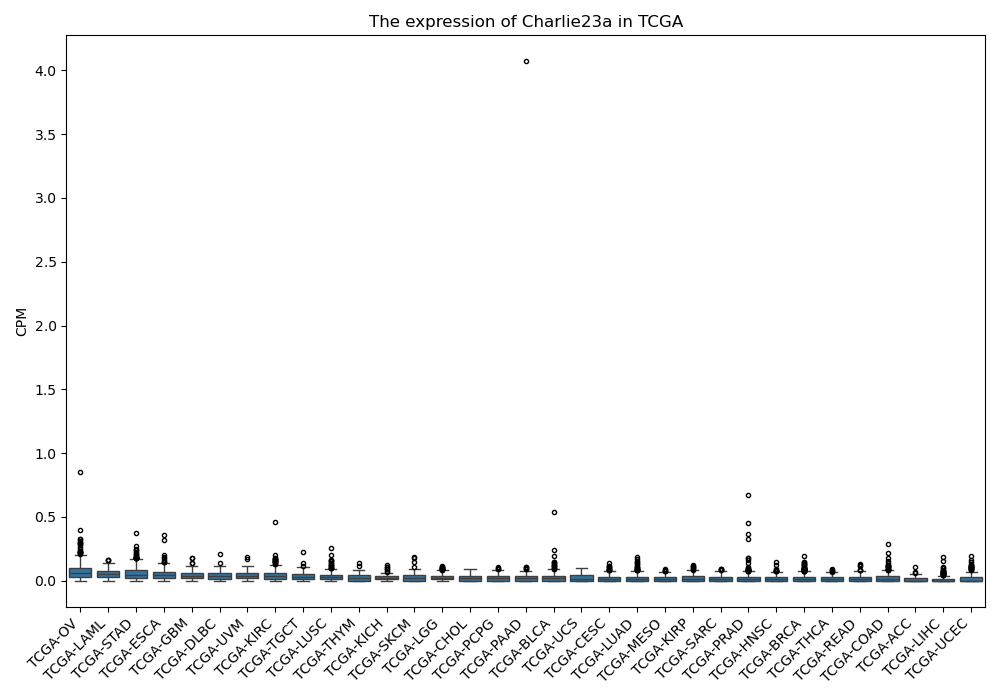
Charlie23a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000100 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 339 |
| Kimura value | 29.77 |
| Tau index | 0.8944 |
| Description | hAT-Charlie DNA transposon, Charlie23a subfamily (non-autonomous) |
| Comment | Charlie23a has 16bp TIRs. The TIR sequence is shared with Charlie18a, and similar to that of Charlie15a and Charlie21a. Present in marsupials; absent from platypus. |
| Sequence |
CAGTGGTTCCCAACCTGTGGGCCGCGGACCCTGGGGGGCTNCGGAGTTATTGCAAGGGGTCCGCGAATCCATATGCGCACCAAGCATTGTCTGTAGACTGAATAAATAATATGTCTCGCACATATATGTACTACTATTTTTCAAATGTATGTAGATGCTGTTGTGATTAATAAAATACAAATTCATTTAAAAGCGTTACTGAATTCATAGTAGCTTTTTGTCATTATGTATTTTCTCAATCAACGGATACTAAAAGTACAACTACTATCATGTGGGGATCCGCGAAATTTTTTGACGTTAAAAAGGGGTCCTCATACTCGAAAAGGTTGGGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie23a | sug | 29 | 40 | - | 12.34 | AGCCCCCCAGGG |
| Charlie23a | MZF1 | 273 | 280 | - | 12.33 | GATCCCCA |
| Charlie23a | MYB30 | 258 | 269 | + | 12.33 | ACAACTACTATC |
| Charlie23a | PBX1 | 235 | 243 | + | 12.28 | CTCAATCAA |
| Charlie23a | ZHD5 | 163 | 170 | + | 12.25 | GTGATTAA |
| Charlie23a | TCX3 | 173 | 184 | + | 12.21 | AAATACAAATTC |
| Charlie23a | rib | 281 | 289 | + | 12.18 | CGCGAAATT |
| Charlie23a | ZNF454 | 19 | 35 | + | 12.17 | GGGCCGCGGACCCTGGG |
| Charlie23a | BtbVII | 225 | 231 | - | 12.17 | ATACATA |
| Charlie23a | mef-2 | 134 | 140 | - | 12.10 | AAAATAG |
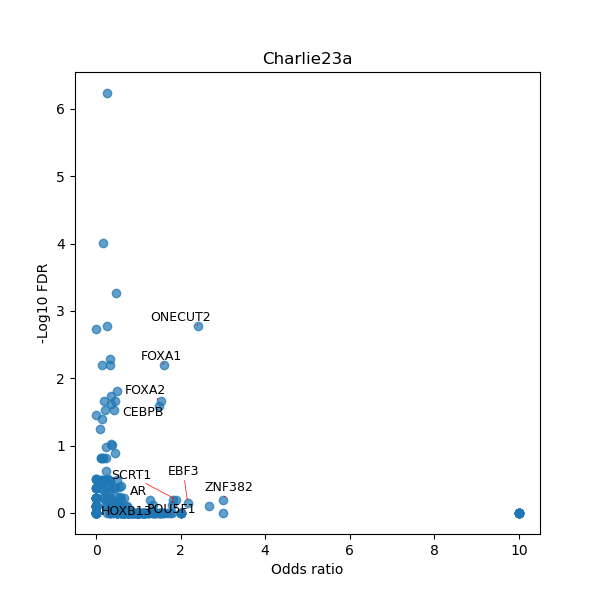
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.