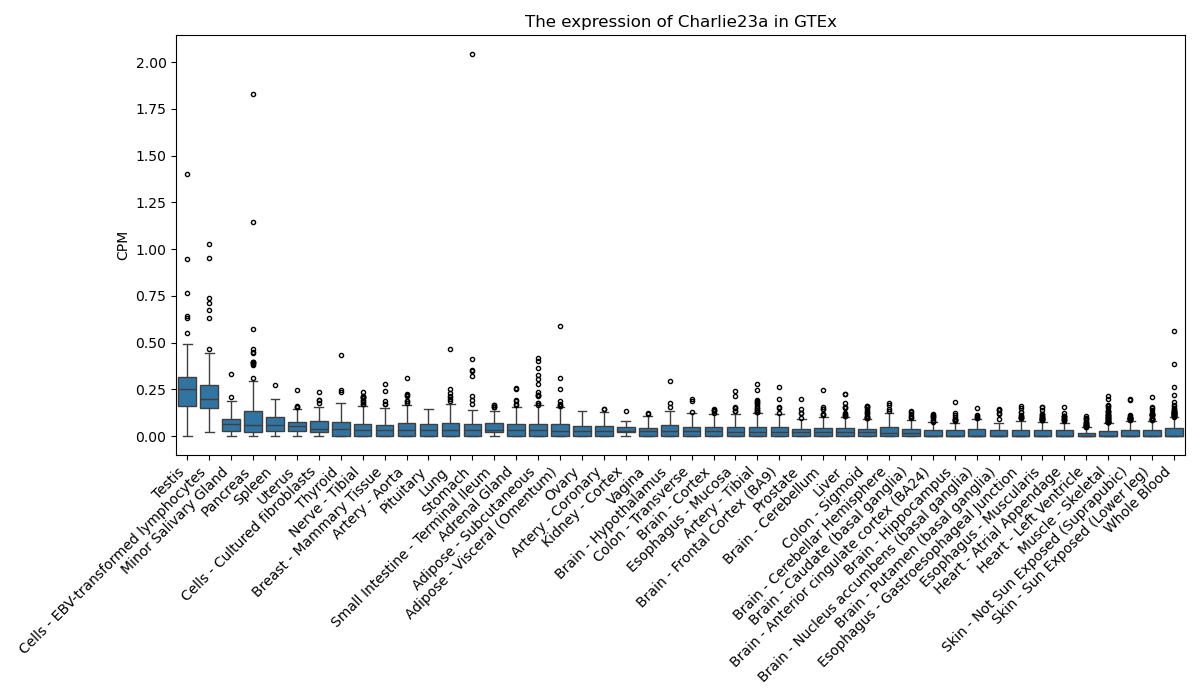
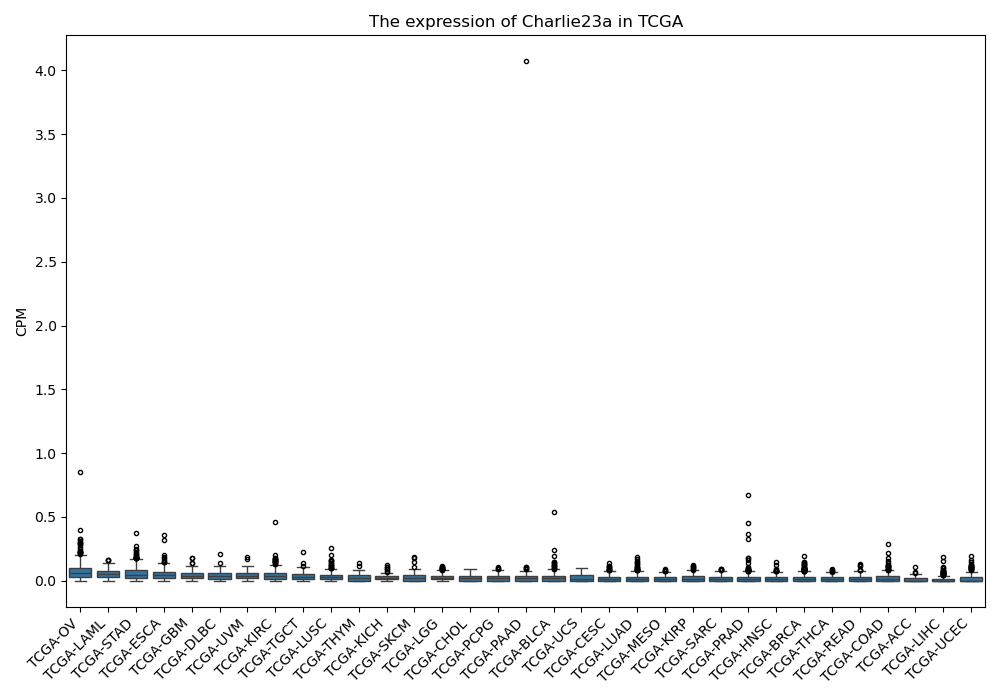
Charlie23a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000100 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 339 |
| Kimura value | 29.77 |
| Tau index | 0.8944 |
| Description | hAT-Charlie DNA transposon, Charlie23a subfamily (non-autonomous) |
| Comment | Charlie23a has 16bp TIRs. The TIR sequence is shared with Charlie18a, and similar to that of Charlie15a and Charlie21a. Present in marsupials; absent from platypus. |
| Sequence |
CAGTGGTTCCCAACCTGTGGGCCGCGGACCCTGGGGGGCTNCGGAGTTATTGCAAGGGGTCCGCGAATCCATATGCGCACCAAGCATTGTCTGTAGACTGAATAAATAATATGTCTCGCACATATATGTACTACTATTTTTCAAATGTATGTAGATGCTGTTGTGATTAATAAAATACAAATTCATTTAAAAGCGTTACTGAATTCATAGTAGCTTTTTGTCATTATGTATTTTCTCAATCAACGGATACTAAAAGTACAACTACTATCATGTGGGGATCCGCGAAATTTTTTGACGTTAAAAAGGGGTCCTCATACTCGAAAAGGTTGGGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie23a | IDD4 | 215 | 229 | - | 17.26 | ACATAATGACAAAAA |
| Charlie23a | IDD6 | 215 | 230 | + | 16.84 | TTTTTGTCATTATGTA |
| Charlie23a | IDD6 | 215 | 230 | + | 16.80 | TTTTTGTCATTATGTA |
| Charlie23a | IDD7 | 216 | 229 | - | 15.48 | ACATAATGACAAAA |
| Charlie23a | POU4F1 | 99 | 110 | + | 14.96 | TGAATAAATAAT |
| Charlie23a | UB3 | 253 | 262 | + | 14.87 | AAAGTACAAC |
| Charlie23a | POU4F3 | 99 | 110 | + | 14.74 | TGAATAAATAAT |
| Charlie23a | IDD5 | 216 | 229 | + | 14.47 | TTTTGTCATTATGT |
| Charlie23a | SBP6 | 254 | 261 | + | 14.34 | AAGTACAA |
| Charlie23a | NRG1 | 26 | 33 | - | 14.28 | CAGGGTCC |
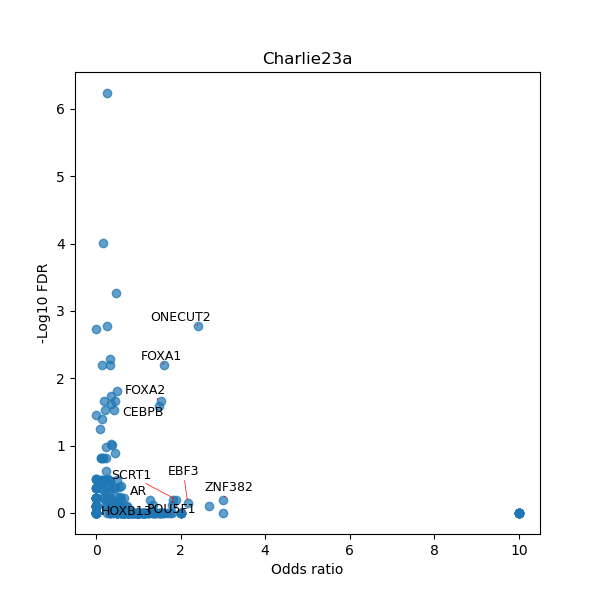
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.