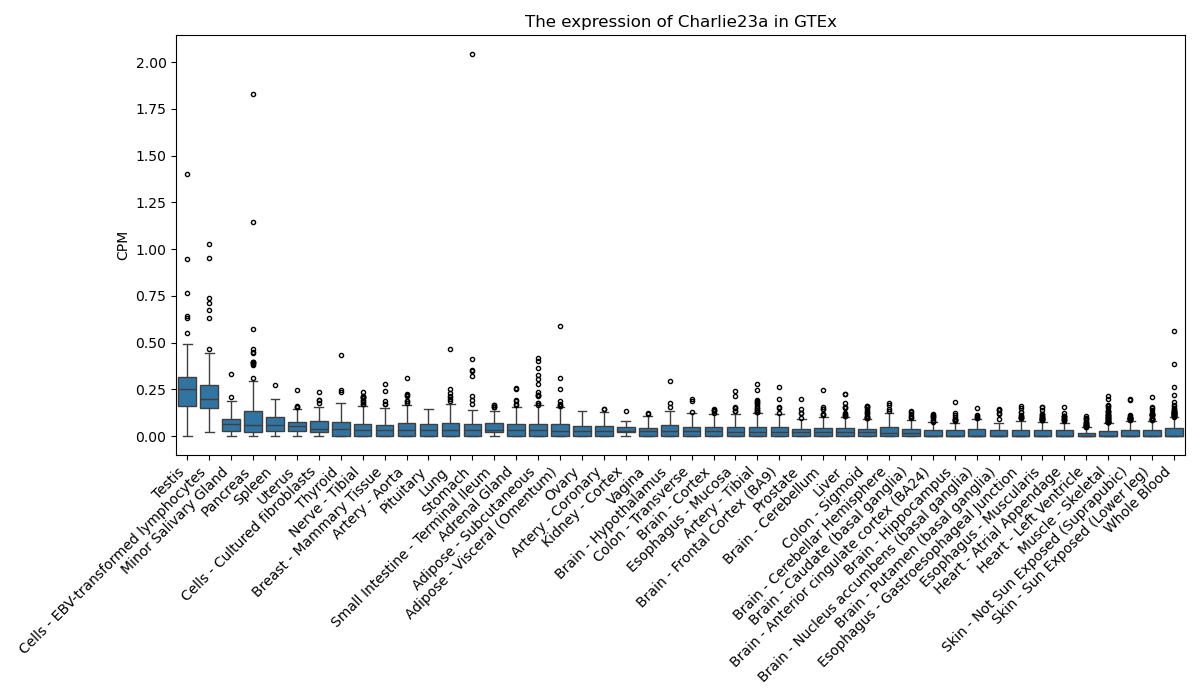
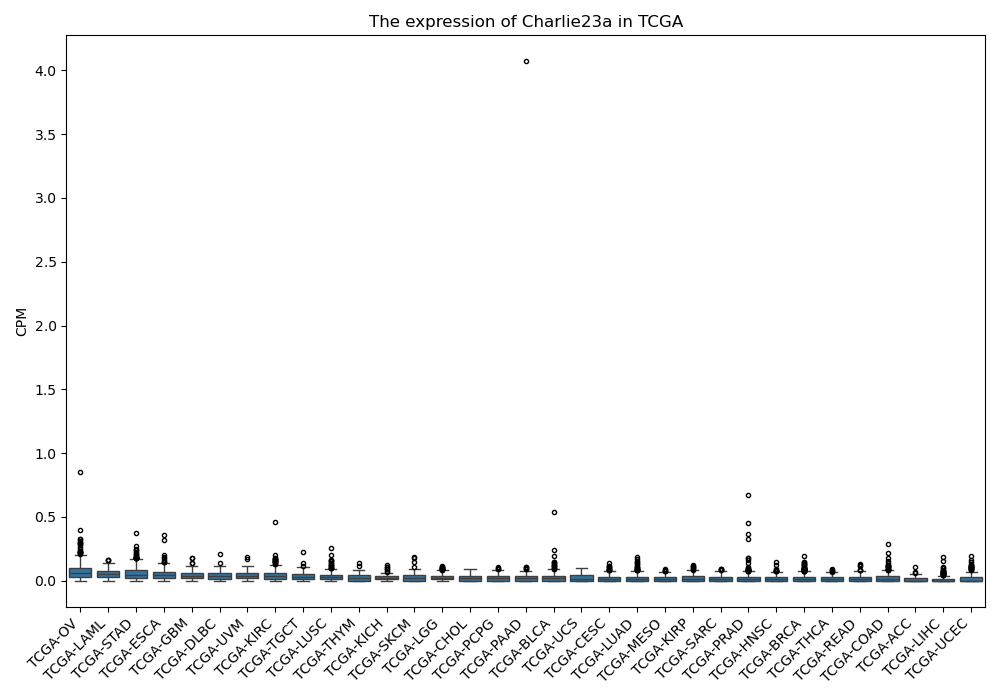
Charlie23a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000100 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 339 |
| Kimura value | 29.77 |
| Tau index | 0.8944 |
| Description | hAT-Charlie DNA transposon, Charlie23a subfamily (non-autonomous) |
| Comment | Charlie23a has 16bp TIRs. The TIR sequence is shared with Charlie18a, and similar to that of Charlie15a and Charlie21a. Present in marsupials; absent from platypus. |
| Sequence |
CAGTGGTTCCCAACCTGTGGGCCGCGGACCCTGGGGGGCTNCGGAGTTATTGCAAGGGGTCCGCGAATCCATATGCGCACCAAGCATTGTCTGTAGACTGAATAAATAATATGTCTCGCACATATATGTACTACTATTTTTCAAATGTATGTAGATGCTGTTGTGATTAATAAAATACAAATTCATTTAAAAGCGTTACTGAATTCATAGTAGCTTTTTGTCATTATGTATTTTCTCAATCAACGGATACTAAAAGTACAACTACTATCATGTGGGGATCCGCGAAATTTTTTGACGTTAAAAAGGGGTCCTCATACTCGAAAAGGTTGGGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie23a | nit-4 | 22 | 28 | - | 14.25 | TCCGCGG |
| Charlie23a | JKD | 216 | 227 | + | 14.05 | TTTTGTCATTAT |
| Charlie23a | SPL8D1 | 125 | 134 | - | 14.02 | GTAGTACATA |
| Charlie23a | SPL8 | 122 | 137 | + | 13.63 | ATATATGTACTACTAT |
| Charlie23a | MGP | 215 | 227 | - | 13.53 | ATAATGACAAAAA |
| Charlie23a | DOF4.5 | 207 | 219 | - | 13.50 | AAAAAGCTACTAT |
| Charlie23a | IDD1 | 216 | 227 | + | 13.33 | TTTTGTCATTAT |
| Charlie23a | IDD11 | 216 | 227 | - | 13.20 | ATAATGACAAAA |
| Charlie23a | SBP8 | 254 | 261 | + | 13.19 | AAGTACAA |
| Charlie23a | URC2 | 42 | 49 | + | 12.98 | CGGAGTTA |
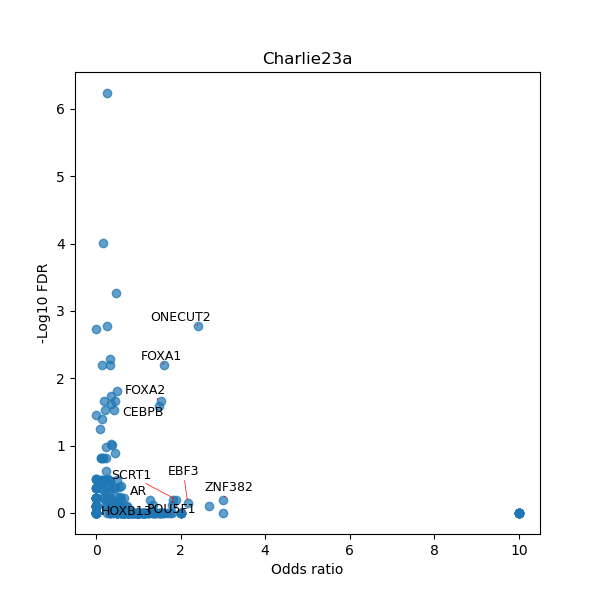
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.