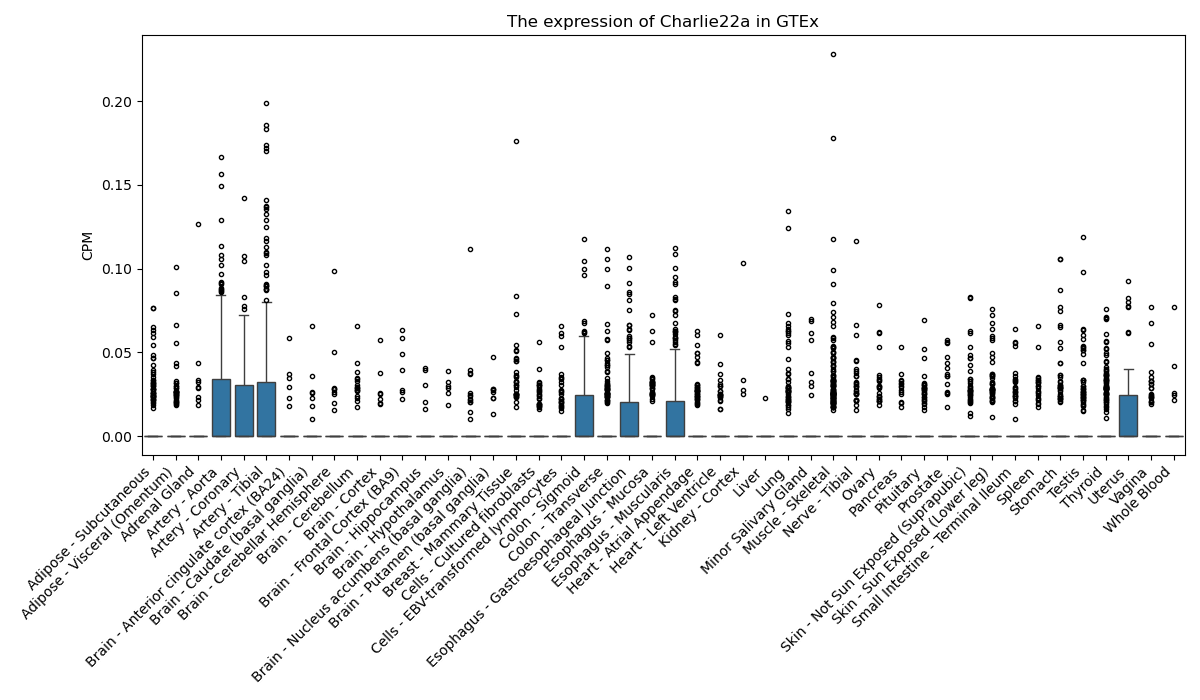
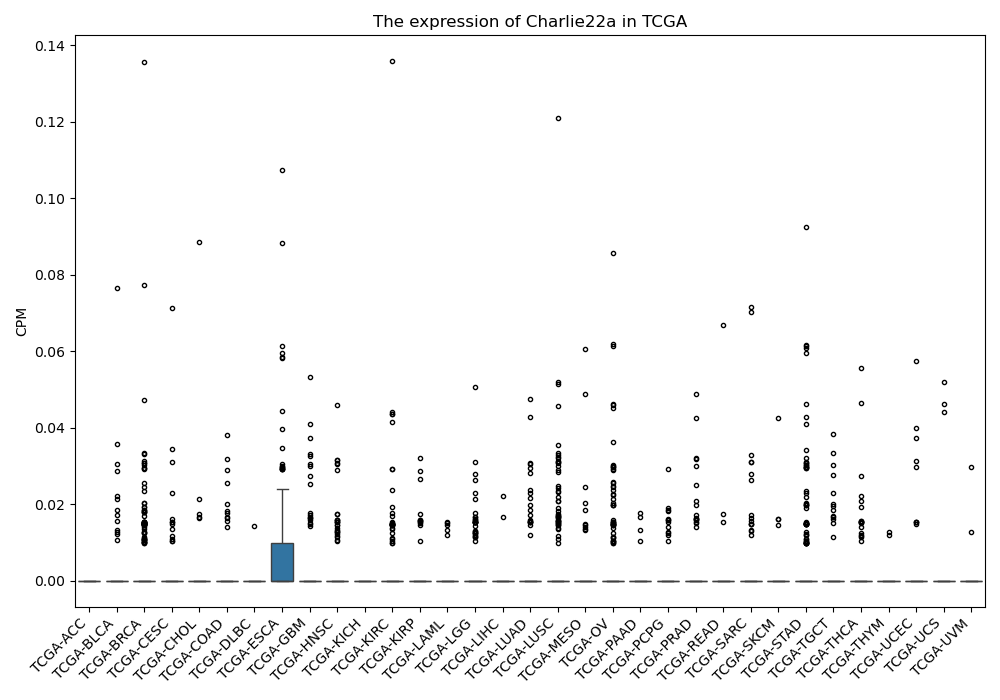
Charlie22a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000099 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 491 |
| Kimura value | 30.03 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Charlie22a subfamily (non-autonomous) |
| Comment | 16 bp TIR; TIRs identical to MER5A. |
| Sequence |
CAGTGGTTCTCAAACTATGCTCCCACGGAACACTGGTGTTCCGCGAGCTGGACCGAGGTGTTCCGCCGCTAAAATCGAATAATGGCAGTCTTTTCCCAATTCGCAGAAAAAATTAAGTTAATGGATAGAATTTTCTCAATTTTAAGTTTTTCTCCCATAAATTTTTCTTAAACCTTTGGCGCCTGCTACCGCTTGTCTGCTAGCGAGCGCTAGCAGATGACAAAACGAATGAACAAGTAGCAAATGAATATTAACGGAAAAACTCGGAAACAGCCGATCATGCTTAGTTCGACAGCTTGTTTTTTTCATGGACACGTGTGCTCGTGGTTGTGATTTTATATTGCATACATATAATATAGTCATATTTTCTGCTAGTAAAATTGTTCCTAGTTCGTATGATGGAATTAAATATATCAGTATTTCATGGTGTTCCGCAAAGAGCACCATGACTTCCAGGTGCTCTGCCACCTGAACAAGTTTGAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie22a | CLOCK | 313 | 319 | - | 12.97 | ACACGTG |
| Charlie22a | blmp-1 | 133 | 143 | - | 12.96 | AAAATTGAGAA |
| Charlie22a | AT5G61620 | 119 | 132 | + | 12.95 | TAATGGATAGAATT |
| Charlie22a | TCF4 | 453 | 460 | - | 12.93 | GCACCTGG |
| Charlie22a | AT2G38300 | 125 | 134 | - | 12.92 | AAAATTCTAT |
| Charlie22a | ZFP14 | 449 | 463 | - | 12.90 | AGAGCACCTGGAAGT |
| Charlie22a | TFAP4::ETV1 | 180 | 192 | - | 12.86 | GCGGTAGCAGGCG |
| Charlie22a | PK18009.1 | 312 | 318 | + | 12.84 | ACACGTG |
| Charlie22a | PK18009.1 | 313 | 319 | - | 12.84 | ACACGTG |
| Charlie22a | HAC1 | 311 | 317 | + | 12.77 | GACACGT |
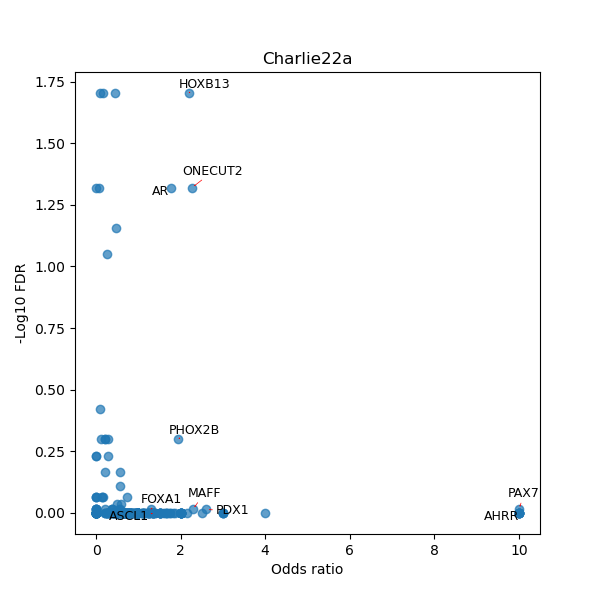
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.