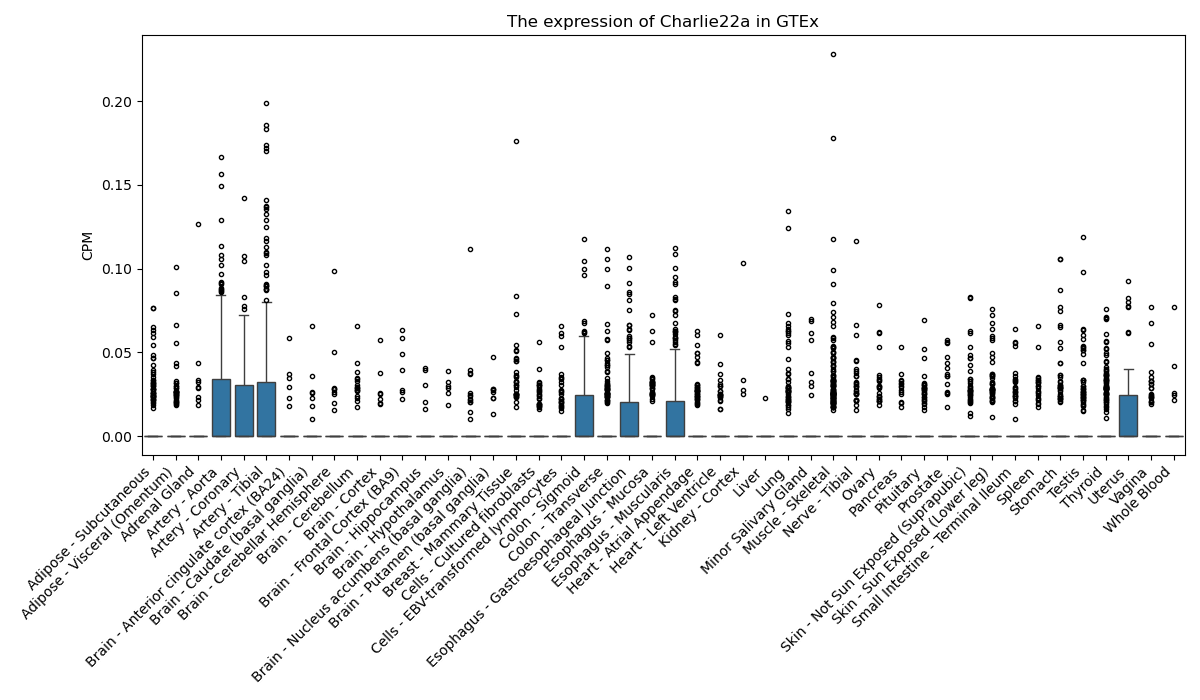
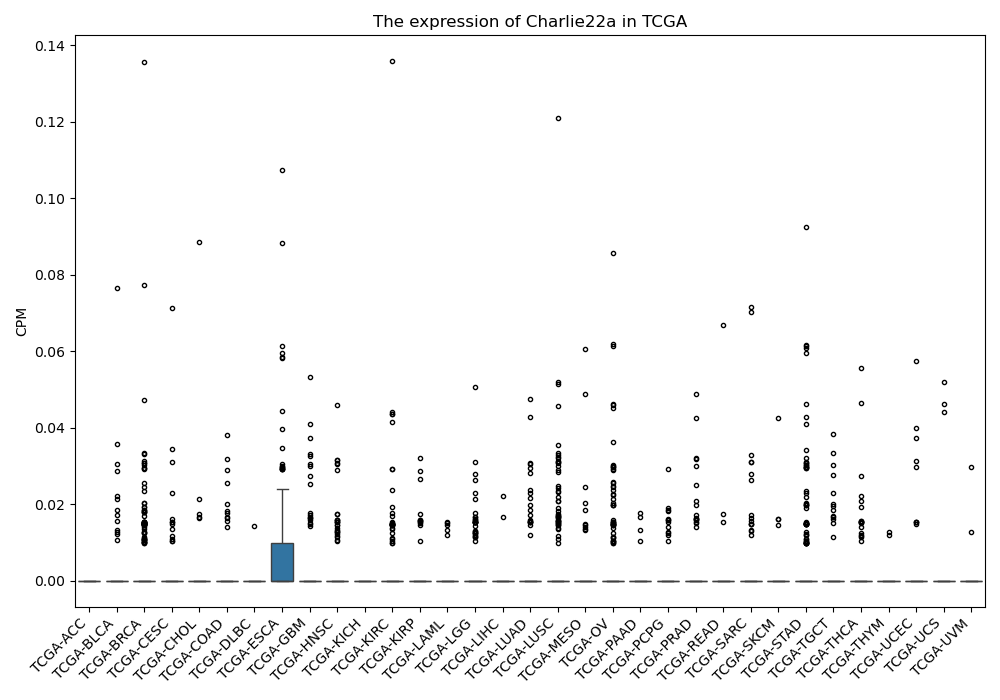
Charlie22a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000099 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 491 |
| Kimura value | 30.03 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Charlie22a subfamily (non-autonomous) |
| Comment | 16 bp TIR; TIRs identical to MER5A. |
| Sequence |
CAGTGGTTCTCAAACTATGCTCCCACGGAACACTGGTGTTCCGCGAGCTGGACCGAGGTGTTCCGCCGCTAAAATCGAATAATGGCAGTCTTTTCCCAATTCGCAGAAAAAATTAAGTTAATGGATAGAATTTTCTCAATTTTAAGTTTTTCTCCCATAAATTTTTCTTAAACCTTTGGCGCCTGCTACCGCTTGTCTGCTAGCGAGCGCTAGCAGATGACAAAACGAATGAACAAGTAGCAAATGAATATTAACGGAAAAACTCGGAAACAGCCGATCATGCTTAGTTCGACAGCTTGTTTTTTTCATGGACACGTGTGCTCGTGGTTGTGATTTTATATTGCATACATATAATATAGTCATATTTTCTGCTAGTAAAATTGTTCCTAGTTCGTATGATGGAATTAAATATATCAGTATTTCATGGTGTTCCGCAAAGAGCACCATGACTTCCAGGTGCTCTGCCACCTGAACAAGTTTGAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie22a | Sox102F | 23 | 34 | - | 13.74 | AGTGTTCCGTGG |
| Charlie22a | ERF118 | 63 | 70 | + | 13.65 | CCGCCGCT |
| Charlie22a | BEH3 | 311 | 320 | + | 13.53 | GACACGTGTG |
| Charlie22a | Stat92E | 98 | 109 | + | 13.50 | AATTCGCAGAAA |
| Charlie22a | DUX4 | 397 | 407 | - | 13.30 | TAATTCCATCA |
| Charlie22a | TFAP4::ETV1 | 264 | 276 | + | 13.28 | TCGGAAACAGCCG |
| Charlie22a | BAM8 | 311 | 319 | + | 13.22 | GACACGTGT |
| Charlie22a | CEP3 | 263 | 269 | + | 13.09 | CTCGGAA |
| Charlie22a | RDR1 | 430 | 436 | - | 13.06 | TGCGGAA |
| Charlie22a | CLOCK | 312 | 318 | + | 12.97 | ACACGTG |
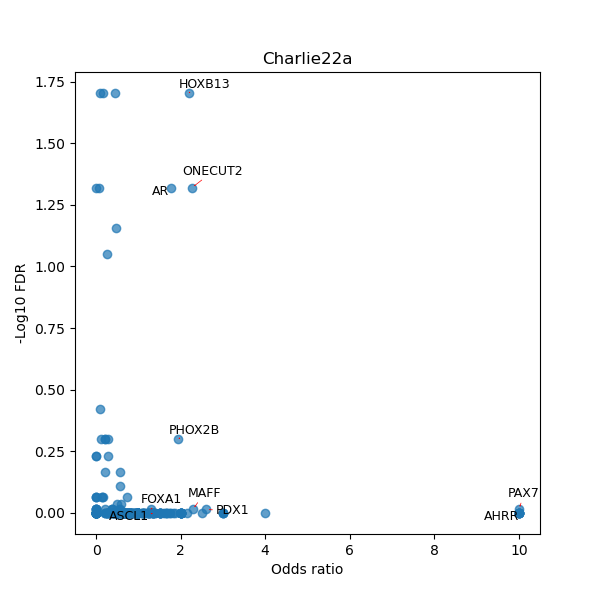
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.