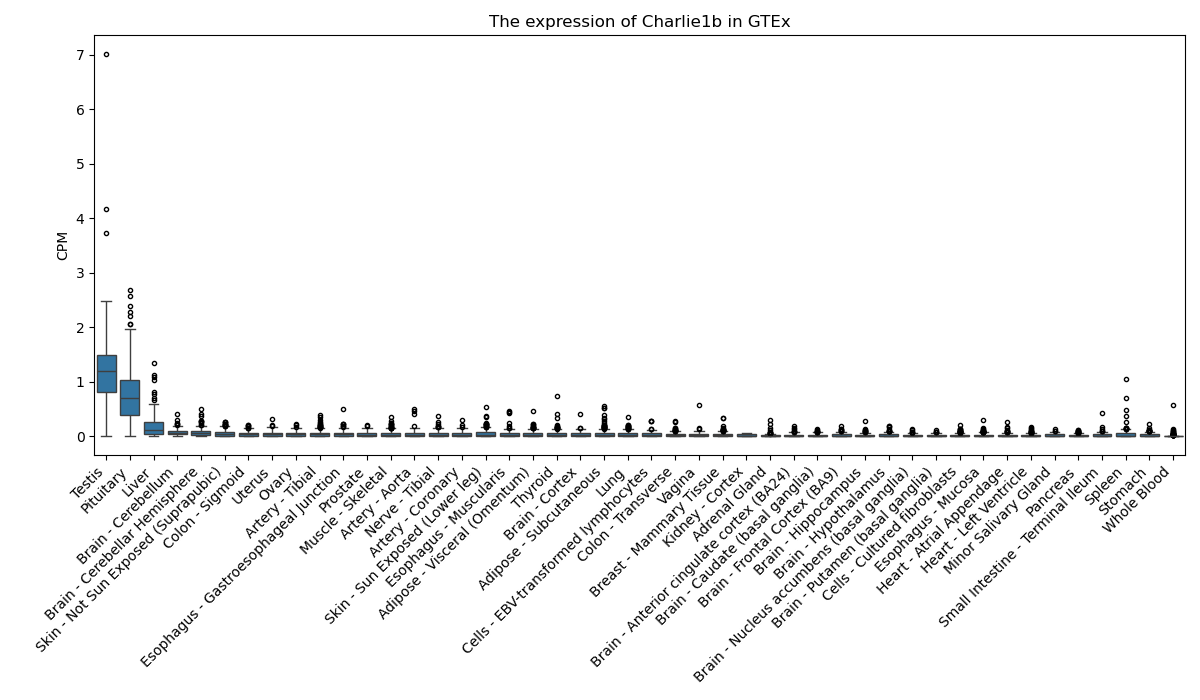
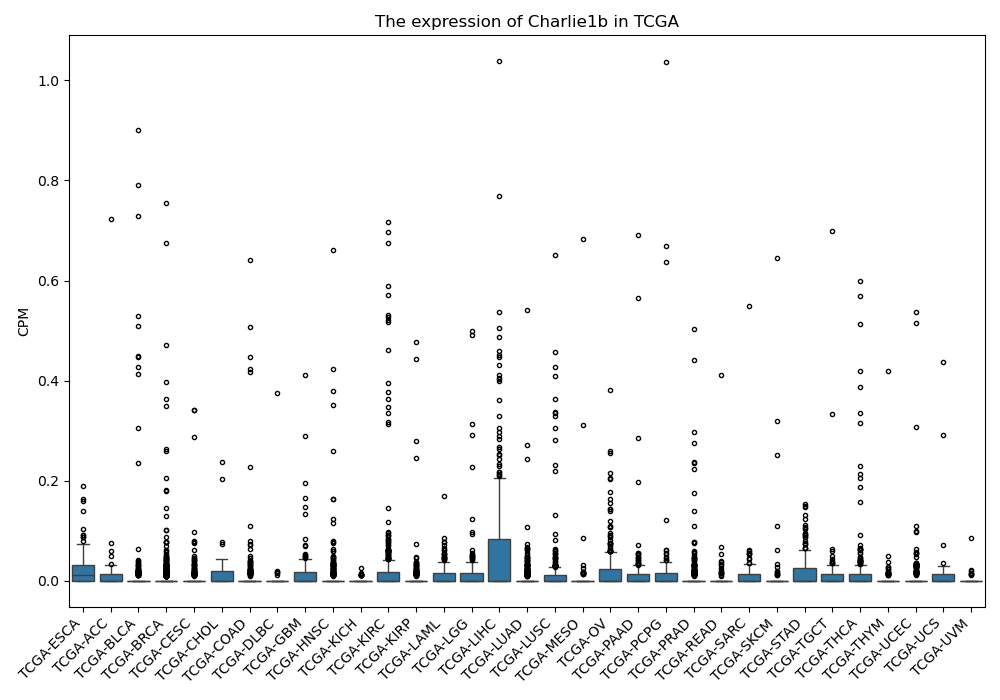
Charlie1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000022 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 523 |
| Kimura value | 20.57 |
| Tau index | 0.9715 |
| Description | hAT-Charlie DNA transposon, Charlie1b subfamily (non-autonomous) |
| Comment | Another internal deletion product of Charlie1. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCCCTGGGGGTCCCCGAGACCCTTTCAGGGGGTCCGCGAGGTCAAAACTATTTTCATAATAATACTAAGACGTTATTTGCCTTTTTCACTCTCATTCTCTCACGAGTGTACAGTGGAGTTTTCCAGAGGCTACATGACGTGTGATATCGCAACAGATTGAATGCAGAAGCAGATATGAGAATCCAGCTGTCTTCTATTAAGCCAGACATTAAAGAGATTTGCAAAAATGTAAAACAATGCCACTCTTCTCACTAAATTTTTTTGTTTTGGAAAATATAGTTATTTTTCATAAAAATATGTTATTTATGTTAACATGTAATGGGTTTATTATTATTTTTAAATGAATTAATAAATATTTTAAAAATTTCTCAGTTTTAATTTCTAATACGGTAAATATCGATAGATATAACCCACATAAACAAAAGCTCTTTGGGGTCCTCAATAATTTTTAAGAGTGTAAAGGGGTCCTGAGACCAAAAAGTTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie1b | blmp-1 | 111 | 121 | - | 13.53 | GAGAGTGAAAA |
| Charlie1b | FOXF2 | 442 | 450 | + | 13.49 | CATAAACAA |
| Charlie1b | EBF1 | 28 | 38 | - | 13.49 | CCCCCAGGGGT |
| Charlie1b | FoxP | 442 | 450 | + | 13.45 | CATAAACAA |
| Charlie1b | VIP1 | 211 | 220 | - | 13.43 | AGACAGCTGG |
| Charlie1b | REI1 | 53 | 59 | - | 13.40 | CCCCTGA |
| Charlie1b | ATHB-51 | 468 | 475 | - | 13.40 | AATTATTG |
| Charlie1b | STAT1::STAT2 | 399 | 411 | + | 13.38 | AGTTTTAATTTCT |
| Charlie1b | ZNF766 | 349 | 357 | - | 13.31 | AATAAACCC |
| Charlie1b | BZIP18 | 211 | 220 | - | 13.31 | AGACAGCTGG |
TFBS enrichment in GRCh38
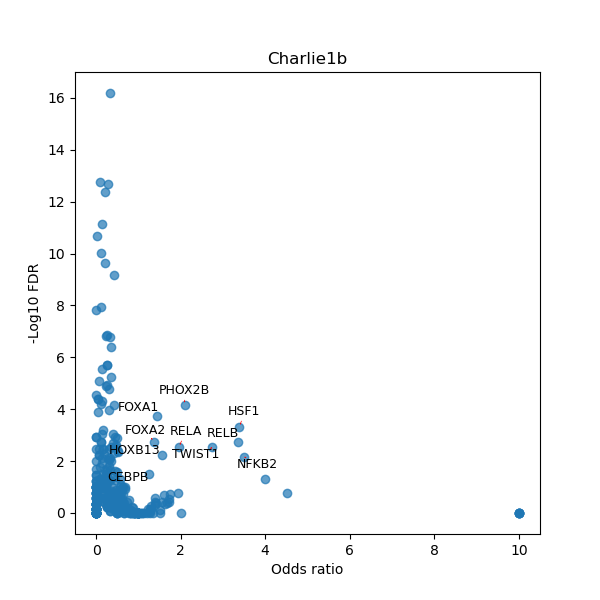
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.