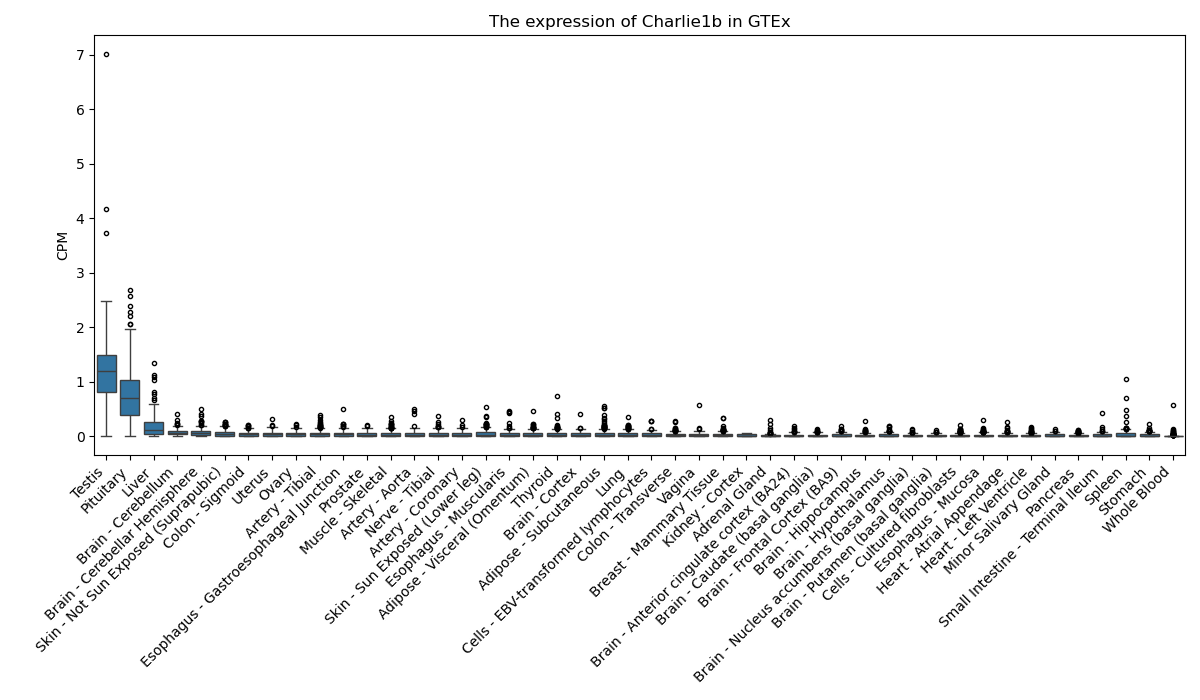
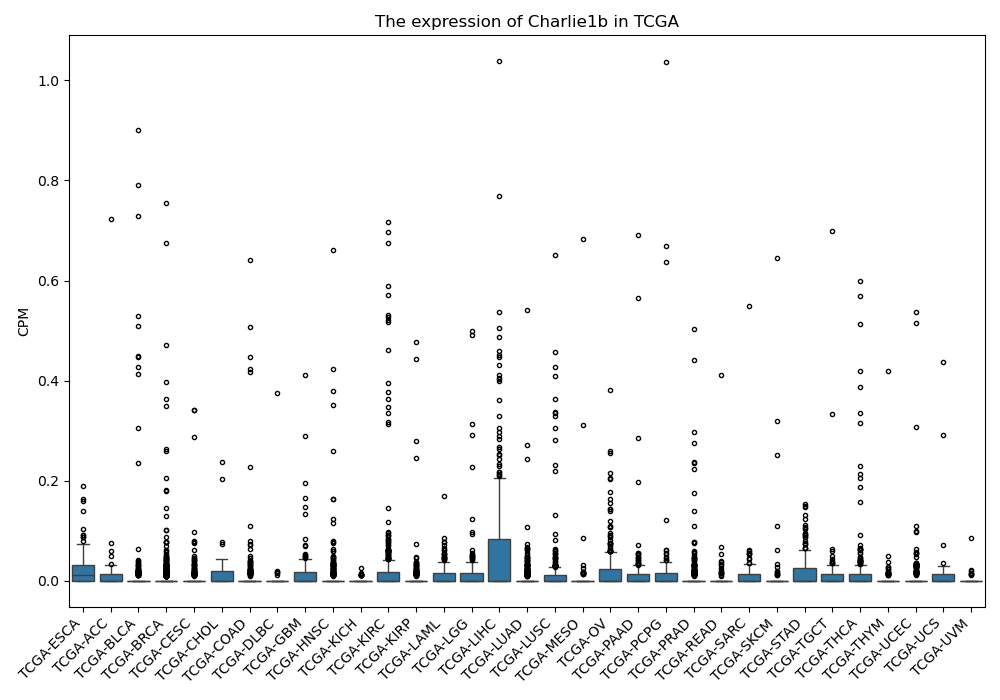
Charlie1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000022 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 523 |
| Kimura value | 20.57 |
| Tau index | 0.9715 |
| Description | hAT-Charlie DNA transposon, Charlie1b subfamily (non-autonomous) |
| Comment | Another internal deletion product of Charlie1. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCCCTGGGGGTCCCCGAGACCCTTTCAGGGGGTCCGCGAGGTCAAAACTATTTTCATAATAATACTAAGACGTTATTTGCCTTTTTCACTCTCATTCTCTCACGAGTGTACAGTGGAGTTTTCCAGAGGCTACATGACGTGTGATATCGCAACAGATTGAATGCAGAAGCAGATATGAGAATCCAGCTGTCTTCTATTAAGCCAGACATTAAAGAGATTTGCAAAAATGTAAAACAATGCCACTCTTCTCACTAAATTTTTTTGTTTTGGAAAATATAGTTATTTTTCATAAAAATATGTTATTTATGTTAACATGTAATGGGTTTATTATTATTTTTAAATGAATTAATAAATATTTTAAAAATTTCTCAGTTTTAATTTCTAATACGGTAAATATCGATAGATATAACCCACATAAACAAAAGCTCTTTGGGGTCCTCAATAATTTTTAAGAGTGTAAAGGGGTCCTGAGACCAAAAAGTTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie1b | Znf423 | 26 | 40 | + | 19.38 | GGACCCCTGGGGGTC |
| Charlie1b | DOF4.2 | 104 | 114 | - | 17.21 | AAAAAGGCAAA |
| Charlie1b | USV1 | 52 | 61 | - | 15.98 | ACCCCCTGAA |
| Charlie1b | BPC1 | 106 | 129 | - | 15.95 | GAGAGAATGAGAGTGAAAAAGGCA |
| Charlie1b | pnr | 423 | 431 | + | 15.64 | TATCGATAG |
| Charlie1b | sug | 29 | 40 | - | 15.13 | GACCCCCAGGGG |
| Charlie1b | Tbx15/18/22 | 165 | 178 | + | 14.66 | ACGTGTGATATCGC |
| Charlie1b | PK27109.1 | 212 | 219 | - | 14.60 | GACAGCTG |
| Charlie1b | BZIP69 | 210 | 220 | - | 14.48 | AGACAGCTGGA |
| Charlie1b | BEAF-32 | 423 | 430 | + | 14.46 | TATCGATA |
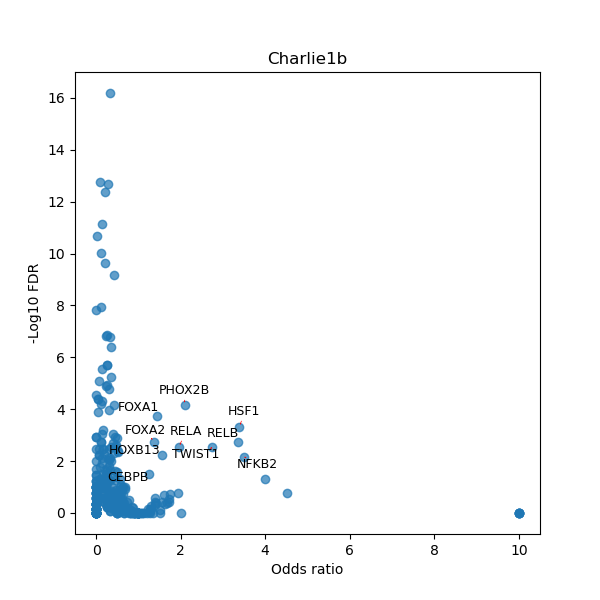
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.