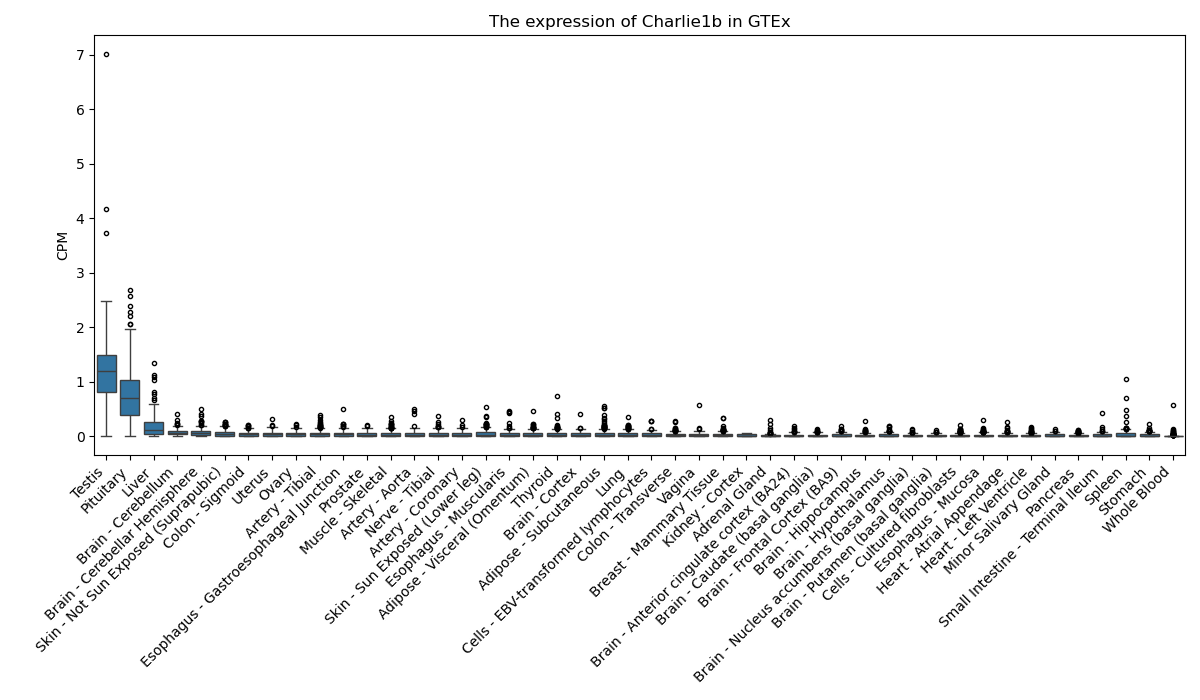
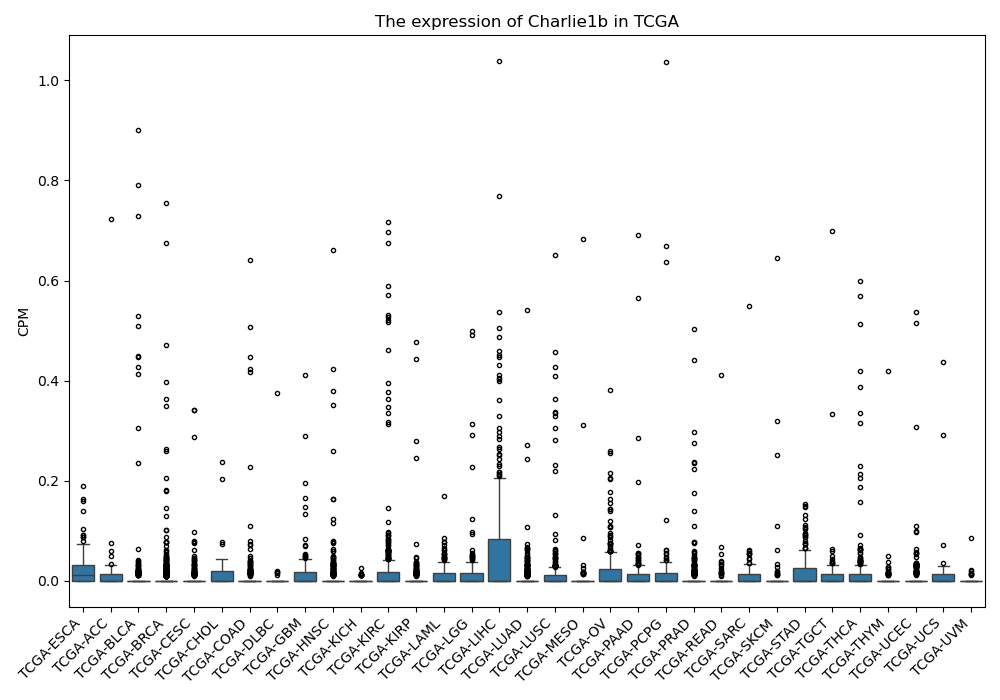
Charlie1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000022 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 523 |
| Kimura value | 20.57 |
| Tau index | 0.9715 |
| Description | hAT-Charlie DNA transposon, Charlie1b subfamily (non-autonomous) |
| Comment | Another internal deletion product of Charlie1. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCCCTGGGGGTCCCCGAGACCCTTTCAGGGGGTCCGCGAGGTCAAAACTATTTTCATAATAATACTAAGACGTTATTTGCCTTTTTCACTCTCATTCTCTCACGAGTGTACAGTGGAGTTTTCCAGAGGCTACATGACGTGTGATATCGCAACAGATTGAATGCAGAAGCAGATATGAGAATCCAGCTGTCTTCTATTAAGCCAGACATTAAAGAGATTTGCAAAAATGTAAAACAATGCCACTCTTCTCACTAAATTTTTTTGTTTTGGAAAATATAGTTATTTTTCATAAAAATATGTTATTTATGTTAACATGTAATGGGTTTATTATTATTTTTAAATGAATTAATAAATATTTTAAAAATTTCTCAGTTTTAATTTCTAATACGGTAAATATCGATAGATATAACCCACATAAACAAAAGCTCTTTGGGGTCCTCAATAATTTTTAAGAGTGTAAAGGGGTCCTGAGACCAAAAAGTTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie1b | let-381 | 443 | 450 | + | 13.99 | ATAAACAA |
| Charlie1b | Sox100B | 335 | 342 | + | 13.99 | TGTTAACA |
| Charlie1b | Sox100B | 335 | 342 | - | 13.99 | TGTTAACA |
| Charlie1b | sex-1 | 66 | 73 | + | 13.83 | GAGGTCAA |
| Charlie1b | ZBTB24 | 491 | 500 | - | 13.77 | CTCAGGACCC |
| Charlie1b | CHES-1-like | 442 | 451 | + | 13.65 | CATAAACAAA |
| Charlie1b | PK00315.1 | 212 | 219 | - | 13.60 | GACAGCTG |
| Charlie1b | Foxj3 | 443 | 451 | + | 13.60 | ATAAACAAA |
| Charlie1b | ATHB-13 | 468 | 475 | + | 13.57 | CAATAATT |
| Charlie1b | Sox6 | 259 | 268 | - | 13.56 | GCATTGTTTT |
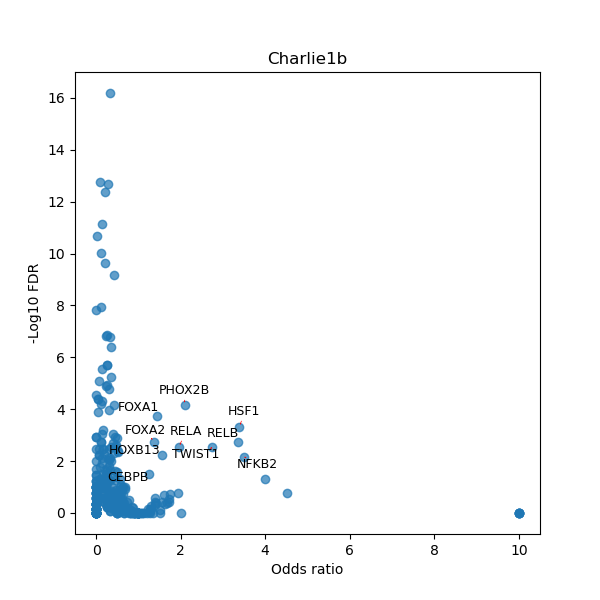
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.