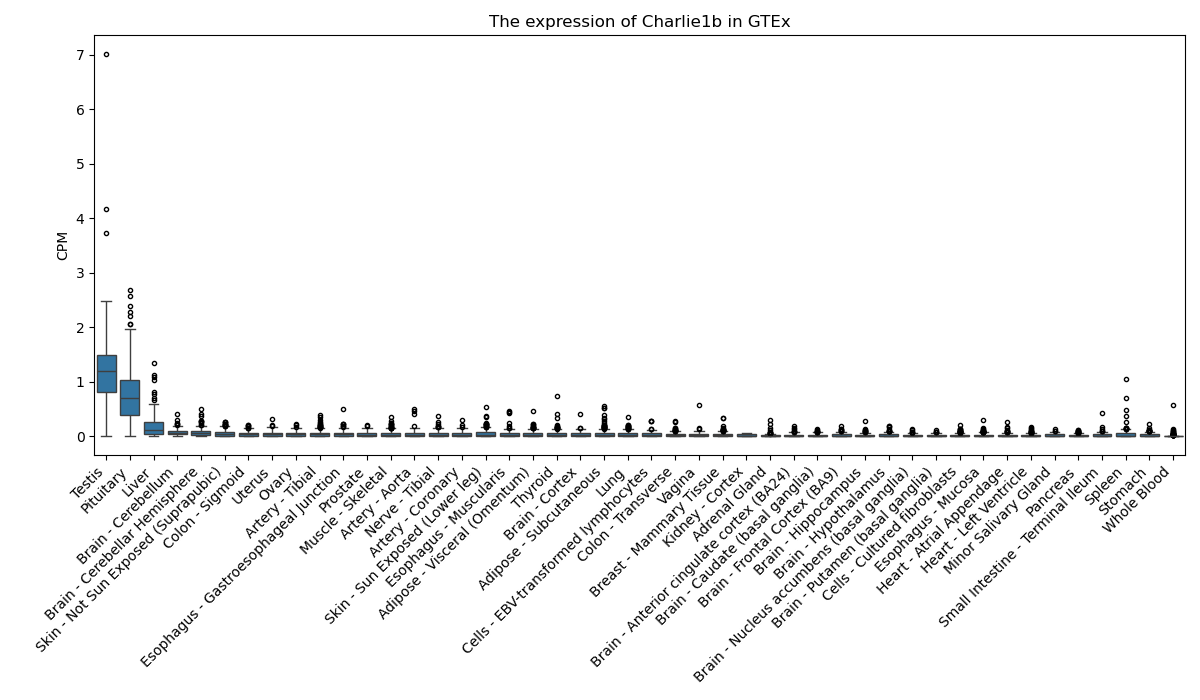
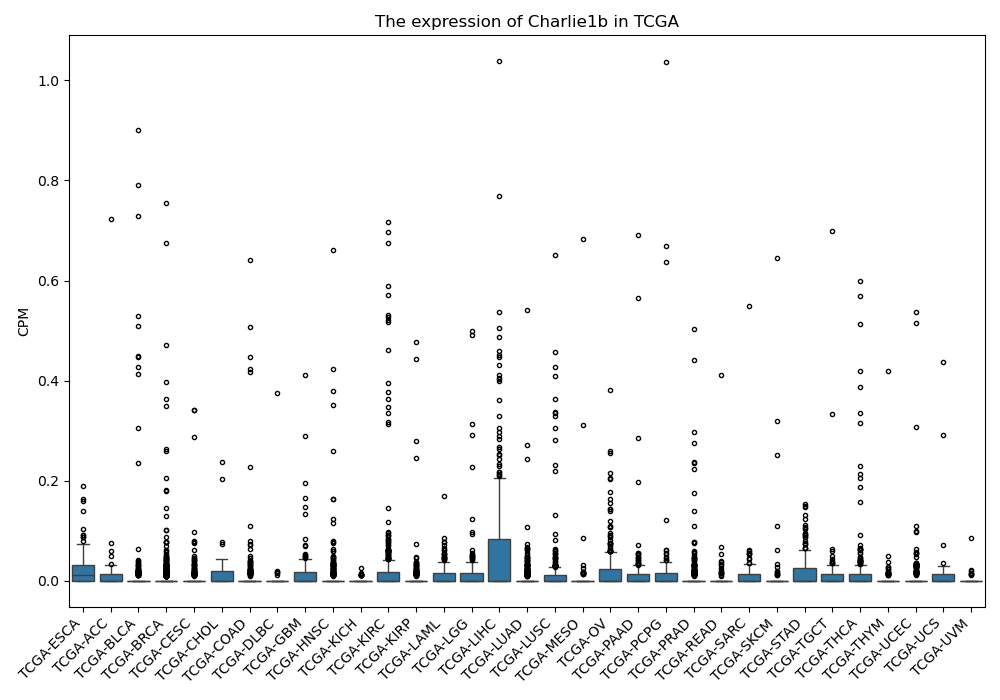
Charlie1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000022 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 523 |
| Kimura value | 20.57 |
| Tau index | 0.9715 |
| Description | hAT-Charlie DNA transposon, Charlie1b subfamily (non-autonomous) |
| Comment | Another internal deletion product of Charlie1. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCCCTGGGGGTCCCCGAGACCCTTTCAGGGGGTCCGCGAGGTCAAAACTATTTTCATAATAATACTAAGACGTTATTTGCCTTTTTCACTCTCATTCTCTCACGAGTGTACAGTGGAGTTTTCCAGAGGCTACATGACGTGTGATATCGCAACAGATTGAATGCAGAAGCAGATATGAGAATCCAGCTGTCTTCTATTAAGCCAGACATTAAAGAGATTTGCAAAAATGTAAAACAATGCCACTCTTCTCACTAAATTTTTTTGTTTTGGAAAATATAGTTATTTTTCATAAAAATATGTTATTTATGTTAACATGTAATGGGTTTATTATTATTTTTAAATGAATTAATAAATATTTTAAAAATTTCTCAGTTTTAATTTCTAATACGGTAAATATCGATAGATATAACCCACATAAACAAAAGCTCTTTGGGGTCCTCAATAATTTTTAAGAGTGTAAAGGGGTCCTGAGACCAAAAAGTTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie1b | BEAF-32 | 423 | 430 | - | 14.46 | TATCGATA |
| Charlie1b | Dref | 423 | 430 | + | 14.38 | TATCGATA |
| Charlie1b | Dref | 423 | 430 | - | 14.38 | TATCGATA |
| Charlie1b | PK16335.1 | 212 | 219 | - | 14.35 | GACAGCTG |
| Charlie1b | PPARG | 33 | 51 | - | 14.21 | AGGGTCTCGGGGACCCCCA |
| Charlie1b | GT-2 | 415 | 425 | - | 14.17 | ATATTTACCGT |
| Charlie1b | ATHB-20 | 468 | 475 | + | 14.11 | CAATAATT |
| Charlie1b | pnr | 422 | 430 | - | 14.05 | TATCGATAT |
| Charlie1b | ceh-39 | 421 | 431 | + | 14.01 | AATATCGATAG |
| Charlie1b | BZIP30 | 211 | 219 | - | 14.00 | GACAGCTGG |
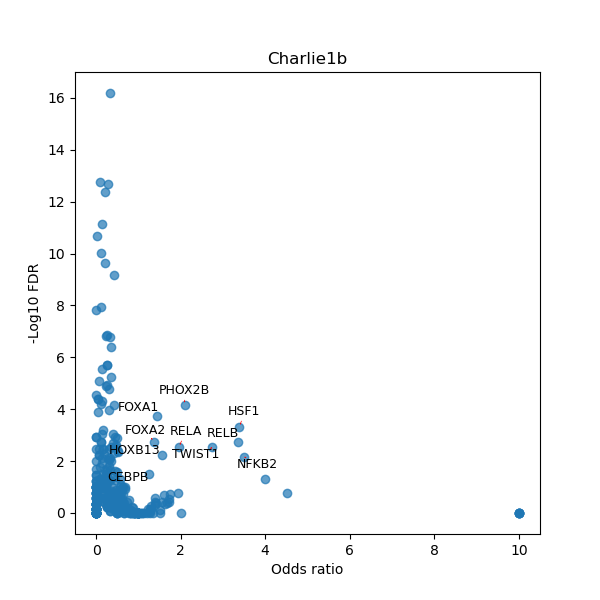
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.