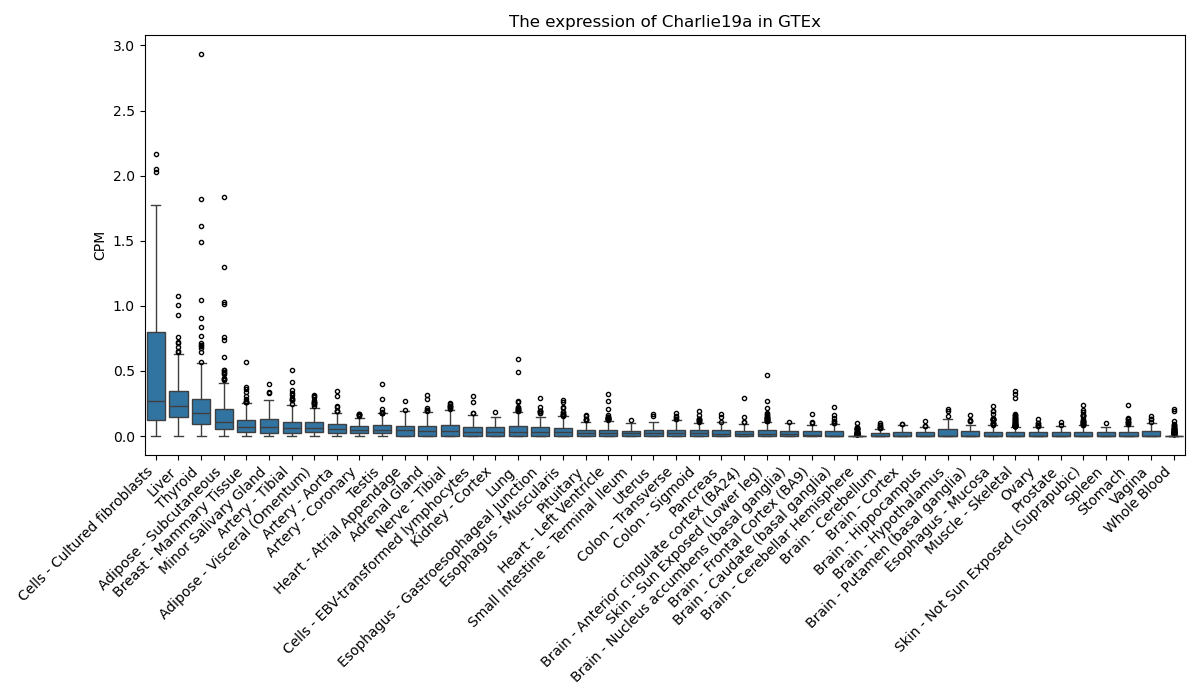
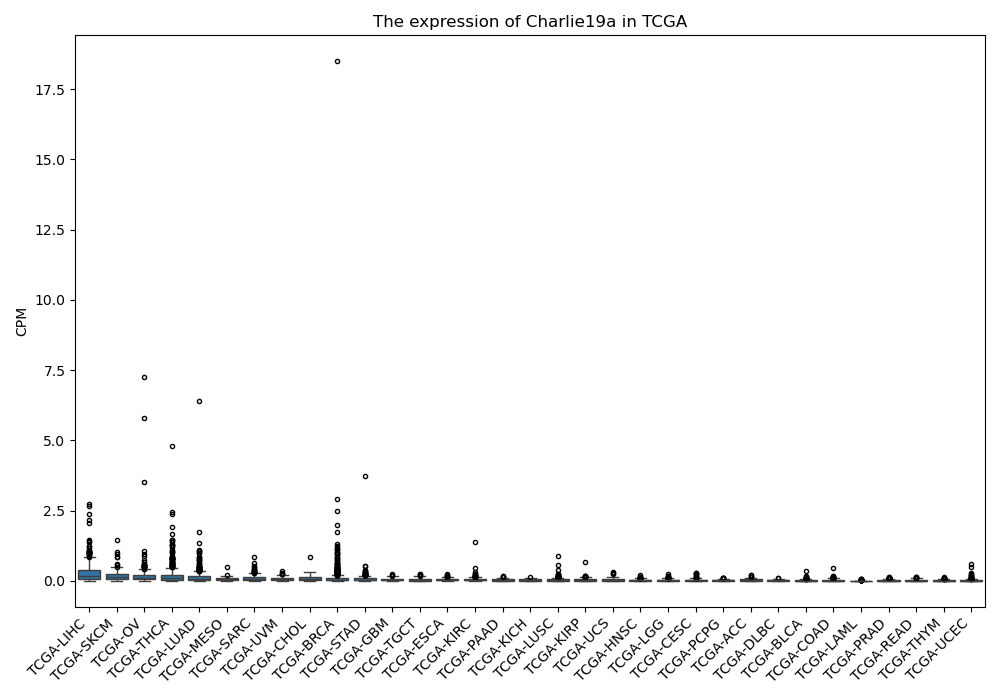
Charlie19a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000096 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 386 |
| Kimura value | 31.34 |
| Tau index | 0.8811 |
| Description | hAT-Charlie DNA transposon, Charlie19a subfamily (non-autonomous) |
| Comment | 16 bp TIRs. |
| Sequence |
CAGTGGTTCCCAACCTGGGGTCCCCGGACCCTAGGGGTCCNTGAAGGTAGTAATGGGGGTCTNCGAGCTATTTTCAATATTTCAAAAAGCCTAACGGAAATCGTACATTTACCCGCGATAAGGCTGCACAGGCTAACTAAAACGTCAAGTTTCTTTGCTTTTGGCTGGAATTATATCAACTCAGTGTGGTAACACTGGTTATCTCATGCTGATCAGAAGCTCTGATTGGCAGTTATATATGTCTTTGATTAATAAAAAATGGGGAAAAAATNAATTATTACTTAATAAATGCAGTTTGTTTGGTAGACAAAATCTTTCAAAACATGGGGTCCATGGGGGAAAATAATAAAAGGGGTCCTTGGTGGTGAAAAGGTTGGGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie19a | Znf423 | 26 | 40 | + | 20.26 | GGACCCTAGGGGTCC |
| Charlie19a | Znf423 | 26 | 40 | - | 18.72 | GGACCCCTAGGGTCC |
| Charlie19a | NTL9 | 277 | 285 | - | 16.85 | TTAAGTAAT |
| Charlie19a | ZNF454 | 19 | 35 | + | 16.26 | GGTCCCCGGACCCTAGG |
| Charlie19a | ceh-60 | 223 | 231 | + | 14.71 | TGATTGGCA |
| Charlie19a | NFYA | 222 | 229 | - | 14.54 | CCAATCAG |
| Charlie19a | M1BP | 187 | 197 | + | 14.49 | TGGTAACACTG |
| Charlie19a | ZHD9 | 246 | 260 | - | 14.48 | ATTTTTTATTAATCA |
| Charlie19a | ZNF454 | 14 | 30 | - | 14.37 | GGTCCGGGGACCCCAGG |
| Charlie19a | ANL2 | 283 | 292 | - | 14.36 | GCATTTATTA |
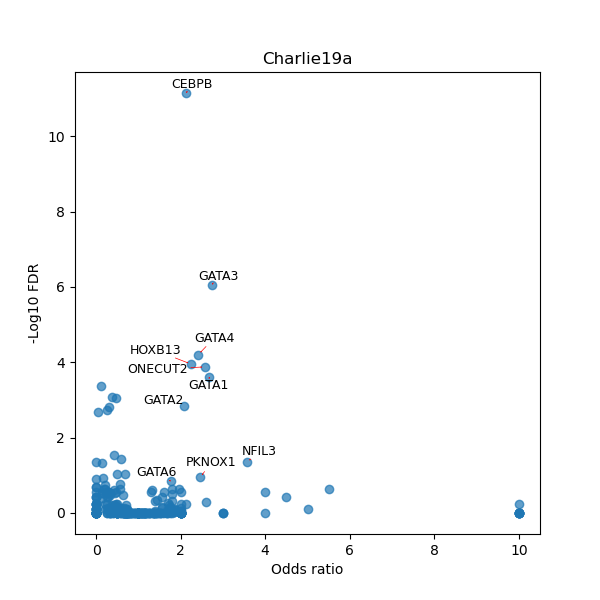
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.