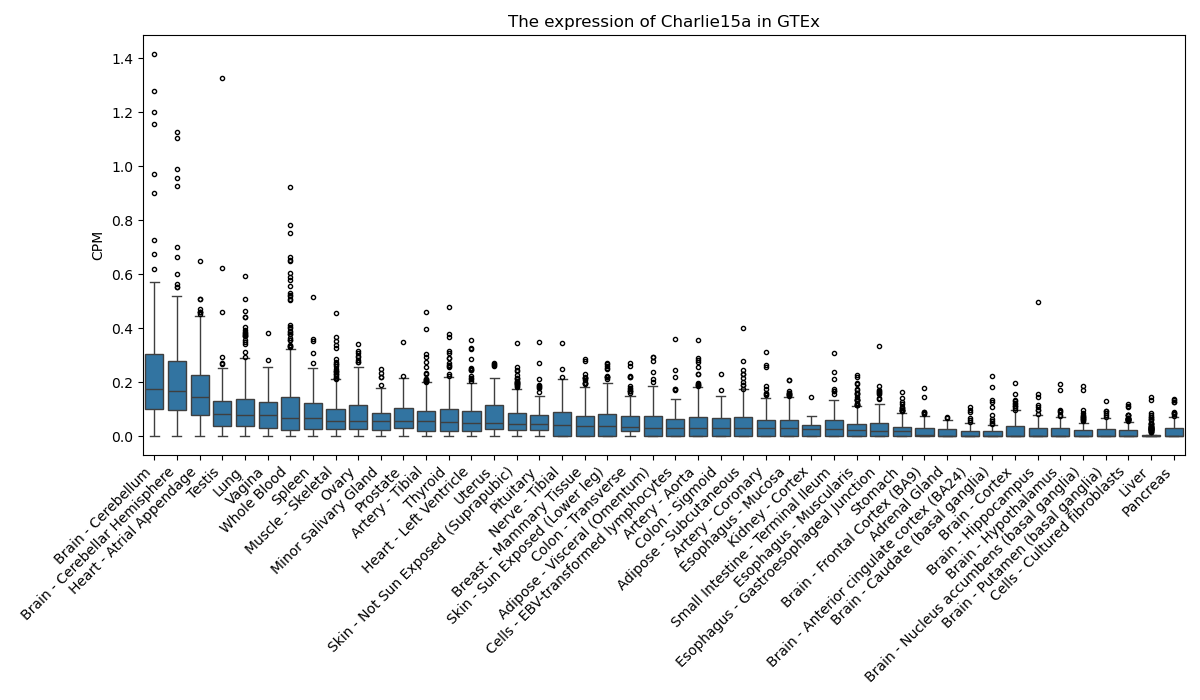
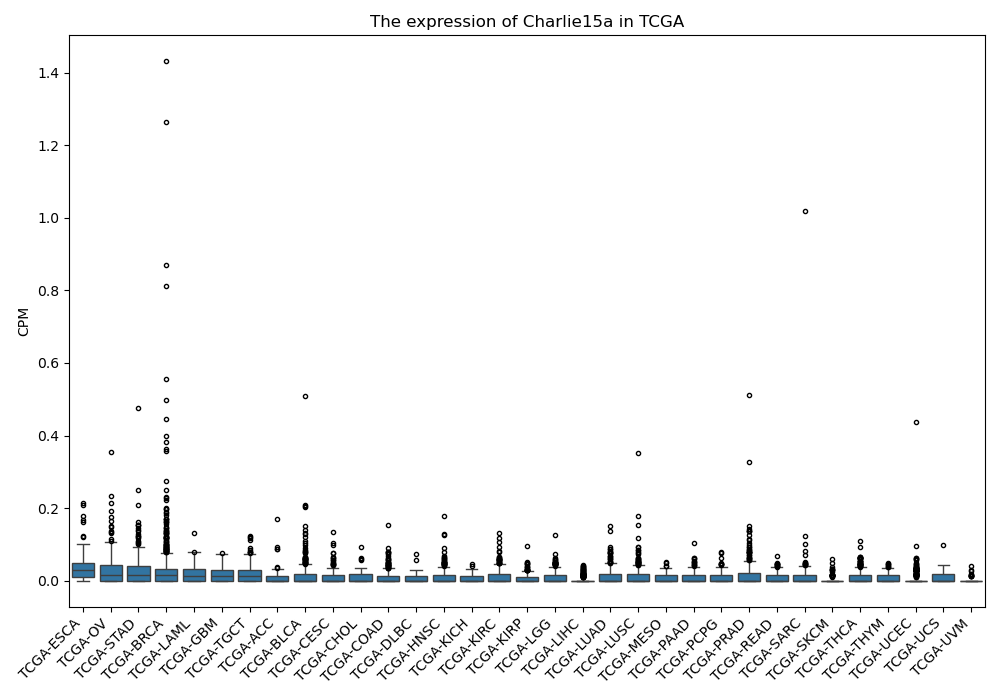
Charlie15a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000089 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 224 |
| Kimura value | 29.28 |
| Tau index | 0.7845 |
| Description | hAT-Charlie DNA transposon, Charlie15a subfamily (non-autonomous) |
| Comment | 8bp Charlie-type TSDs; 16 bp TIRs. |
| Sequence |
CAGTGGTTTTCAAACTGTGTTCCGCGGAGCCCTAGGGGTTCCGCGGAGGTGCCTCGGGGGCTGCTGGGNGGGNNGTGAGGCTGGGCGGGCGGGGCTCTGGGCCTCCCACCCCCGCTTCAACCAGAGCAGCTCCGCTTTTATCTGTTTTATATATTGGGCTTCCGCGTAAGATTTCATTTGAAGAAAGGGTTCTGCTGCTTAAAAAAAAAGTTTGAAAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie15a | FEZF2 | 78 | 85 | - | 15.03 | CCCAGCCT |
| Charlie15a | Klf6-7-like | 80 | 99 | - | 14.93 | CAGAGCCCCGCCCGCCCAGC |
| Charlie15a | KLF15 | 83 | 90 | - | 14.65 | GCCCGCCC |
| Charlie15a | IME1 | 22 | 29 | + | 14.59 | CCGCGGAG |
| Charlie15a | IME1 | 41 | 48 | + | 14.59 | CCGCGGAG |
| Charlie15a | Spps | 85 | 95 | - | 14.44 | GCCCCGCCCGC |
| Charlie15a | ZNF454 | 87 | 103 | - | 14.31 | GGCCCAGAGCCCCGCCC |
| Charlie15a | nit-4 | 21 | 27 | + | 14.25 | TCCGCGG |
| Charlie15a | nit-4 | 22 | 28 | - | 14.25 | TCCGCGG |
| Charlie15a | nit-4 | 40 | 46 | + | 14.25 | TCCGCGG |
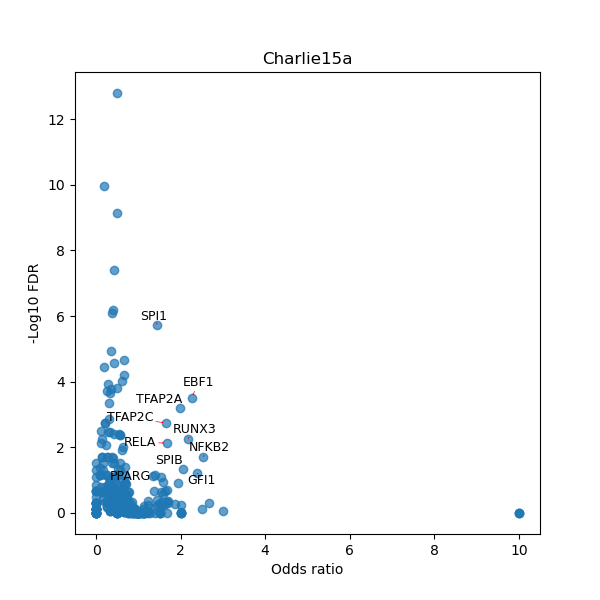
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.