Charlie14a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000088 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 1166 |
| Kimura value | 29.13 |
| Tau index | 0.9891 |
| Description | hAT-Charlie DNA transposon, Charlie14a subfamily |
| Comment | Charlie14a has 16 bp TIRs that closely resemble those of Charlie1. Otherwise the similarity at the DNA level is small. Contains the beginning and end of the coding region. Subfamilies exist with some additional coding sequence, but no full, autonomous element has been detected yet. |
| Sequence |
CAGCATTTCCCAAACTGTGTTCCGTAGAACACTAGTGTTCCGTGAGATGTTAATAGGTGTTCTGTGAAAAAAATCGATGCTAGCGATATTTTTTCTTCTAAAATGTATTTTAAAATATATGTAAGAAAAATACTAAACACATTTATATAGTTNTTATTATTATGCGATAATCTGCCTTTAAGAAATGGTGTGCCATACCCGAGAACGCCAGGAAGTCGCATGAGAGATCACCTGATGCATGAACGCGTGACGTTCGATCTGCGCATTGATGCANAGGCAGCATTTACCAACTAACCATCGCGTTGATGTATGTCGTCTATTCGATGATCGCTGGTCTATGTTATGTATCTTGTCTTATAATTTCAACATATATTTGTGATATTATTCAATGTTTATTGTATATTAAACATTTATACTACACAAACTCATTCCCCATGGATCTGTGGCTGAAAGAAGGAAGTTTTAAACGAAAAGCAGAGTCTTGTGGTTCTCAAACTGTGTCTGAATCAAACAAGCAAAAAGTCGGAATACTTCAAAGTGAAGATGAACAATCGCAGCCTTCGGTTTCTAAAAAATCGGGAGGTGAGCTGTCCAAGATGAAACAATTAATGATTTTTGGAGCAGCCTAATTCAAGAGTACCCAAATATTGCAAAACGTGCAGTTTGTGTTCTTCCCTTTGCTACAACGTACCTGTGCGAAACGGGATTTTCATATTATGCTGCAATGAAAACAAAATATAGGAATAGACTTGATGCTGCACCTGATATGAGGATCCGACTCAGCAACATCATTCCTAATATTAAGCGGATATGTGATATAAAGAATAAAAAGANCGTNCACTGTTCTCATTTTAAAATATTTTTGGTGTATGTCATATTTTATAACAAAAATGTAAATATANATTGCTTTATCTTGNTATATTTTGTTCTTTTTAATNTNAATTAAAATGATTTAATTTCTCTATTTTNANTTCTGTTATAAAGTTAAATAGCATATGAATCAAATTTTGATCATTTCTTGCAGCAATTAATTCATCTGTGAGACCATTTTTTGCTGTCTACTTTAATCCAAGCGCGCATTATATCTCATGAGCTCCTGTTGGTGTTCCGTCAATACATTATAATTTTAAAAGTGTTCCGTGGTCAAATAAGTTTGGGAAATGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie14a | SREBF1 | 227 | 236 | + | 17.14 | ATCACCTGAT |
| Charlie14a | Hmx2 | 1022 | 1036 | + | 16.82 | CAGCAATTAATTCAT |
| Charlie14a | Hmx2 | 600 | 614 | + | 16.79 | AAACAATTAATGATT |
| Charlie14a | ZKSCAN5 | 579 | 587 | + | 16.46 | GGAGGTGAG |
| Charlie14a | NAC071 | 529 | 543 | - | 16.05 | CTTCACTTTGAAGTA |
| Charlie14a | FOXB1 | 890 | 900 | + | 15.56 | AATGTAAATAT |
| Charlie14a | Hmx1 | 1023 | 1031 | + | 15.52 | AGCAATTAA |
| Charlie14a | ATMYB31 | 281 | 295 | + | 15.33 | CATTTACCAACTAAC |
| Charlie14a | MYB31 | 281 | 295 | + | 15.33 | CATTTACCAACTAAC |
| Charlie14a | Hmx3 | 1023 | 1031 | + | 15.27 | AGCAATTAA |
TFBS enrichment in GRCh38
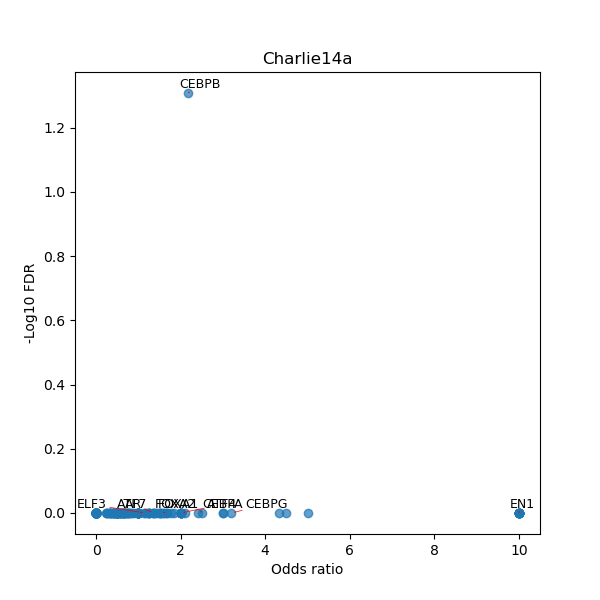
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.