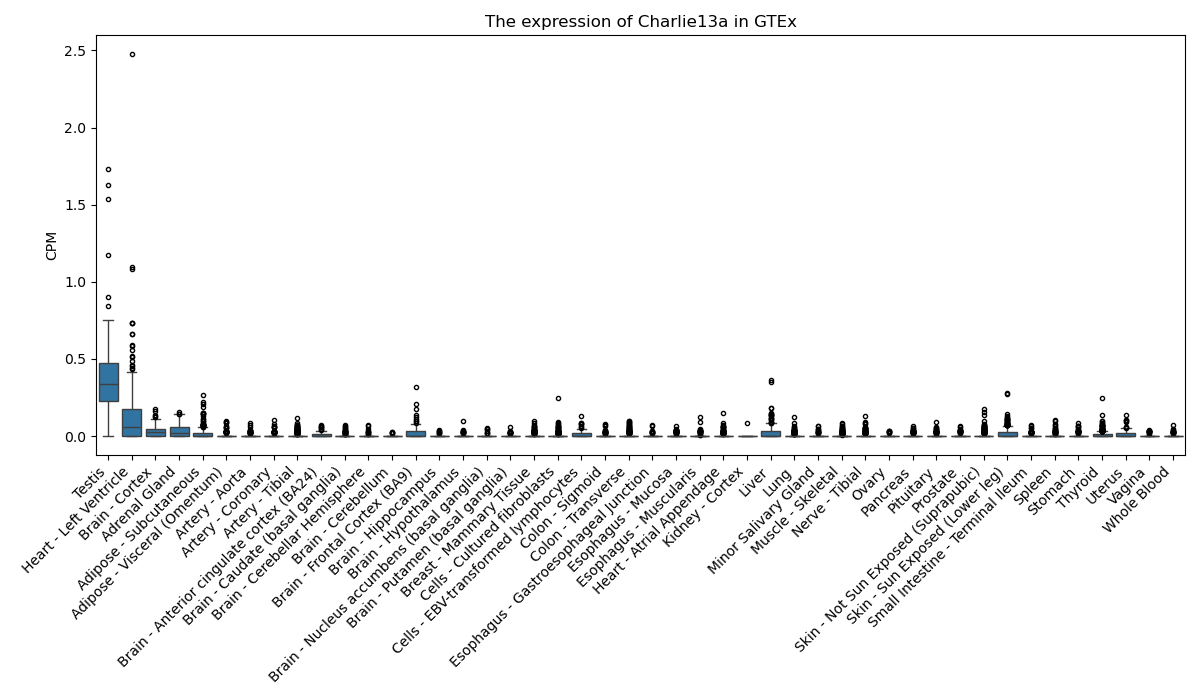
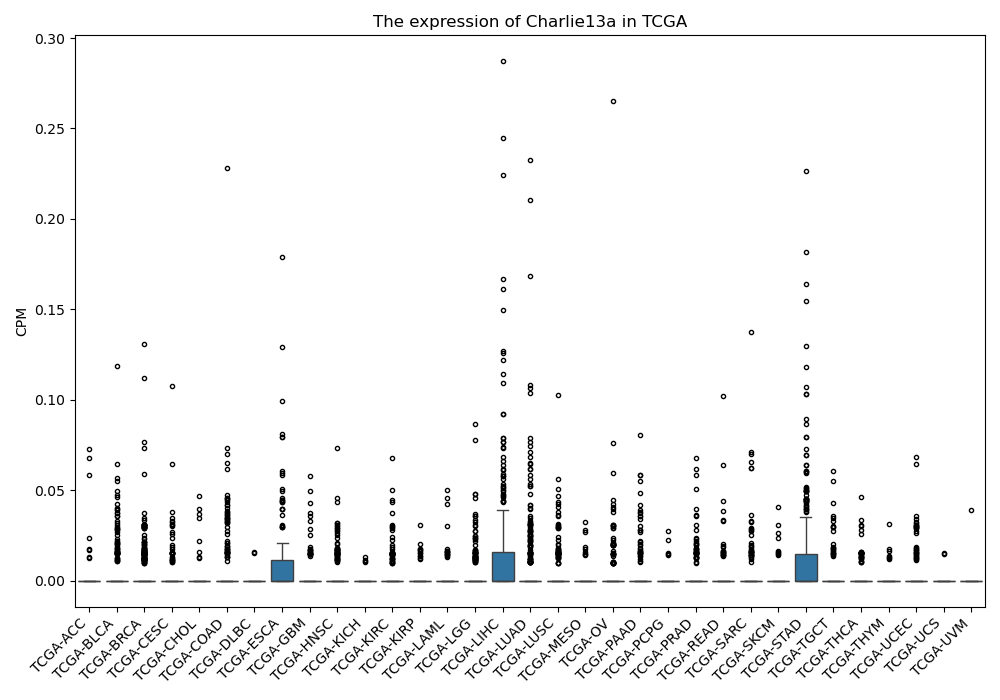
Charlie13a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000086 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Mammalia |
| Length | 1514 |
| Kimura value | 32.78 |
| Tau index | 0.9930 |
| Description | hAT-Charlie DNA transposon, Charlie13a subfamily |
| Comment | - |
| Sequence |
CAGGGACATCCAATATGCAGCCCGCGGGCCAAATGCGGCCCGCCTGACCCCAGGGTGCGGCCCGCCTGAGATTTTTAGCAAAAATGTTTTTACCTAATTAGCTAGCACACATGAACTAGTAGGGCCTAACCACCTGCCAGTAAGGAATTTGAGAAGTTTGGTCTAAACATGGAAAAGCTGAGTTCACTTGCCACAGATGGGGCACCTGCAATGAAAGGGAAGAAAGGTTTCAACAGTTTATTCAAGTCAAAAGAGAAGGTTGGTGTGCCCAGCTTCCATTGCATCATTCATCAGGAGAGTTTGTGCTGTGAACTTTGCAAGTCAGGTTCATTGCATGATGTGATGGAGGAAGTGATCAAGATTGTGAATTTTCTTCGTGCTCGTGCCCTTAACCATCGCCAGTTTATGGGATATTTGGAAGAAGTTGAAGCAGAGTATGGAGATTTAGTTTATTTTAATGCAGTCAGATGGCTAGGTCGTGGAAACTTGTTGAAGAGATTTACTGAGCTGCTTCCTCAGATCAAGGACTTTTTGGAATTAAAGGGCTCTGAGAGACCCAACTTGTCGGACCCCCAGTGGCTCACAAGATTGTACTTTCTTACTGATGTGACAAGACATCTGAACACACTGAACTTGAAACTGCAGGGAAAAAACAAGAGCATTGCTGATTTGTTCAAGGAAGTGCAGGTATTCAGACTCAAACTGGACATGTGGATTGAGCAGATGGCAACTGGAGACTGCACTCATTTCCCATTGCTGAACTGCCCTGAGCTCAATGGAATCACAGATTTTGAAGAACTGCAGAGCTATTTGGTGGAAATGAAGTCACAATTTCAGAAGAGATTCACAGATTTTGATGAGTATGAATCATGCTTTAAGTTTCTGCGGCTGCCTTTGGAGTGCAAACCTCAGGATGTGTCTGAATTATCAAGCATCTGCCCTATCAGTGTGGCTCAGTTTCAAGAGGAACTGATTGAGCTGAAAGCGAACTATGCCCACCAAGAAGACACAGTATCCCTGGAATTTTGGAGATGAGTATATGAATCCCAGAACTATCCTGAAATTTCTGGCTATGTGGCCAAACTCTGGGCAATGTTTGGGTCCACCTGGCTCTGTGAAAGTACATTCTCTTTCATGAAACTGCTGAAATCGAAGCTGAGAGCCACCATCAGTGATGTAAACCTGGAGTCAGAATTGAGATGTGCCCTGACTGAATACACCCCACAGTTCTCGAGAATAGTTCAGTCTGTTAAATACCAGTATTCTCATTGACTTGATCAAAATTGATCAATGACTAGCACTAGGCCTATAACTGTGCACTTTTGTATGAATAAAGTTGATTAAACTGTTTTTTGGACTGCTTTTGTATTACTGCTNAAAATCCATATTTTTGAATAGCTAACAGGCTATATTTTCACCATTGTTATCAATAAAAAAATTCATGCGGCCCGCACACATATGGATTTCTGATCATGTGGCCCACTATTGAAAAAACNTGGACGCCTCTGCTCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie13a | ZmbZIP57 | 335 | 344 | + | 14.61 | ATGATGTGAT |
| Charlie13a | Sox17 | 1418 | 1427 | - | 14.61 | ATAACAATGG |
| Charlie13a | NEUROD1 | 193 | 200 | + | 14.58 | ACAGATGG |
| Charlie13a | Sox5 | 1418 | 1425 | - | 14.38 | AACAATGG |
| Charlie13a | MYB43 | 256 | 267 | - | 14.34 | CACACCAACCTT |
| Charlie13a | SBP6 | 589 | 596 | - | 14.34 | AAGTACAA |
| Charlie13a | sug | 568 | 579 | + | 14.28 | GACCCCCAGTGG |
| Charlie13a | AT3G10030 | 1246 | 1253 | - | 14.25 | TTAACAGA |
| Charlie13a | Elf5 | 676 | 683 | + | 14.24 | AAGGAAGT |
| Charlie13a | ETV5::DRGX | 347 | 358 | + | 14.21 | AGGAAGTGATCA |
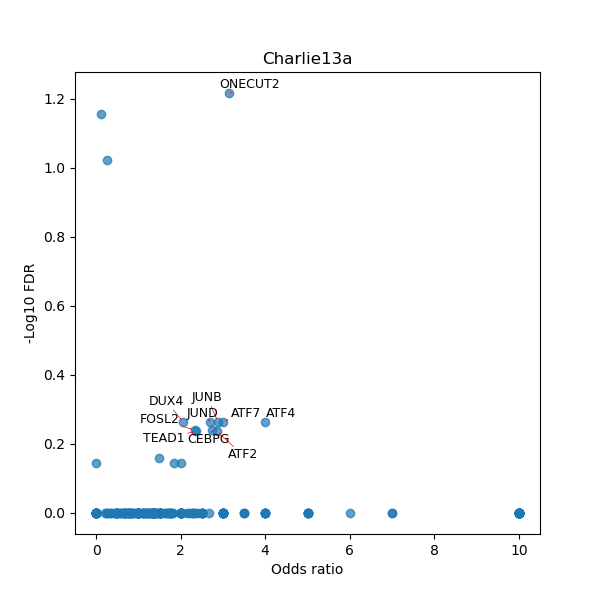
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.