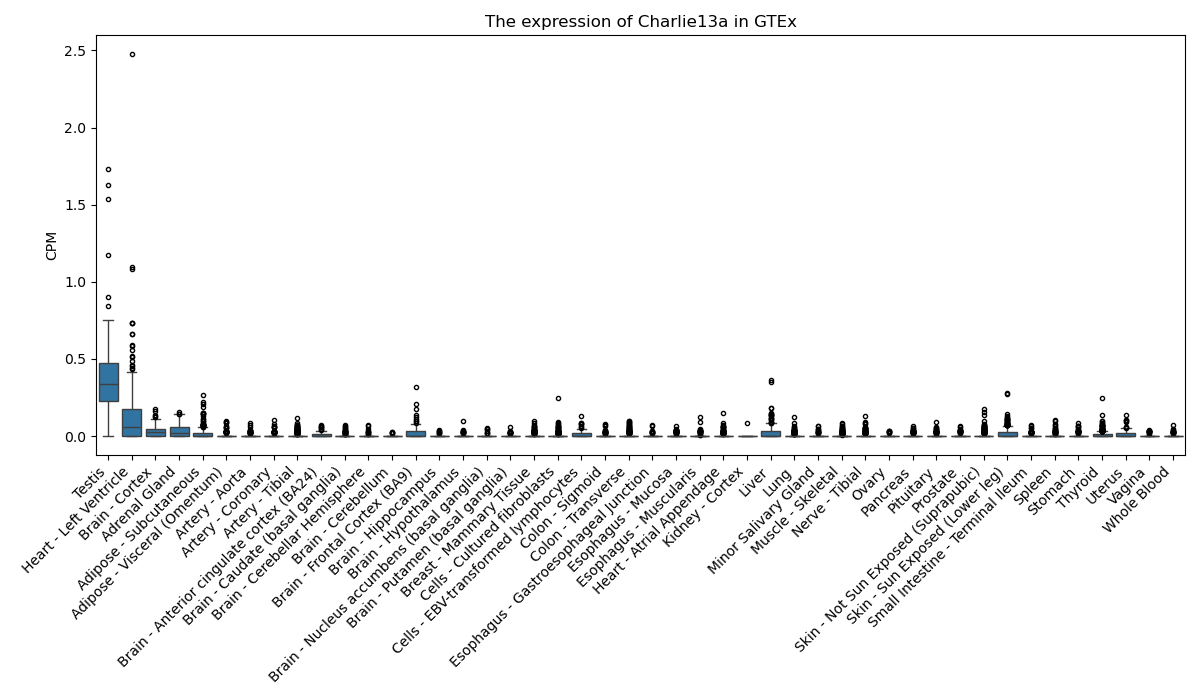
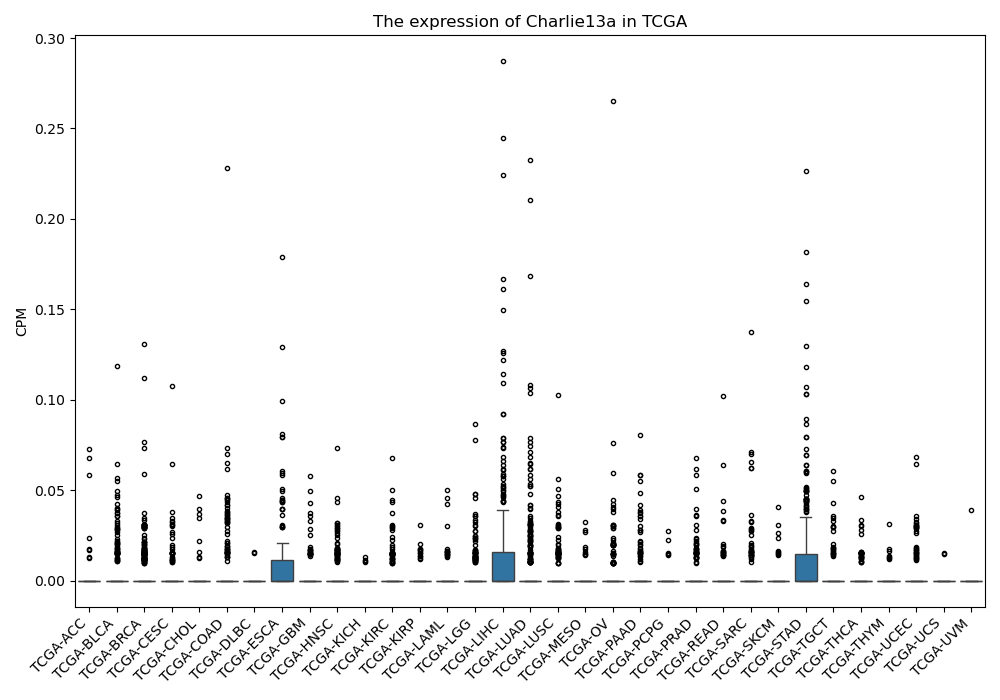
Charlie13a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000086 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Mammalia |
| Length | 1514 |
| Kimura value | 32.78 |
| Tau index | 0.9930 |
| Description | hAT-Charlie DNA transposon, Charlie13a subfamily |
| Comment | - |
| Sequence |
CAGGGACATCCAATATGCAGCCCGCGGGCCAAATGCGGCCCGCCTGACCCCAGGGTGCGGCCCGCCTGAGATTTTTAGCAAAAATGTTTTTACCTAATTAGCTAGCACACATGAACTAGTAGGGCCTAACCACCTGCCAGTAAGGAATTTGAGAAGTTTGGTCTAAACATGGAAAAGCTGAGTTCACTTGCCACAGATGGGGCACCTGCAATGAAAGGGAAGAAAGGTTTCAACAGTTTATTCAAGTCAAAAGAGAAGGTTGGTGTGCCCAGCTTCCATTGCATCATTCATCAGGAGAGTTTGTGCTGTGAACTTTGCAAGTCAGGTTCATTGCATGATGTGATGGAGGAAGTGATCAAGATTGTGAATTTTCTTCGTGCTCGTGCCCTTAACCATCGCCAGTTTATGGGATATTTGGAAGAAGTTGAAGCAGAGTATGGAGATTTAGTTTATTTTAATGCAGTCAGATGGCTAGGTCGTGGAAACTTGTTGAAGAGATTTACTGAGCTGCTTCCTCAGATCAAGGACTTTTTGGAATTAAAGGGCTCTGAGAGACCCAACTTGTCGGACCCCCAGTGGCTCACAAGATTGTACTTTCTTACTGATGTGACAAGACATCTGAACACACTGAACTTGAAACTGCAGGGAAAAAACAAGAGCATTGCTGATTTGTTCAAGGAAGTGCAGGTATTCAGACTCAAACTGGACATGTGGATTGAGCAGATGGCAACTGGAGACTGCACTCATTTCCCATTGCTGAACTGCCCTGAGCTCAATGGAATCACAGATTTTGAAGAACTGCAGAGCTATTTGGTGGAAATGAAGTCACAATTTCAGAAGAGATTCACAGATTTTGATGAGTATGAATCATGCTTTAAGTTTCTGCGGCTGCCTTTGGAGTGCAAACCTCAGGATGTGTCTGAATTATCAAGCATCTGCCCTATCAGTGTGGCTCAGTTTCAAGAGGAACTGATTGAGCTGAAAGCGAACTATGCCCACCAAGAAGACACAGTATCCCTGGAATTTTGGAGATGAGTATATGAATCCCAGAACTATCCTGAAATTTCTGGCTATGTGGCCAAACTCTGGGCAATGTTTGGGTCCACCTGGCTCTGTGAAAGTACATTCTCTTTCATGAAACTGCTGAAATCGAAGCTGAGAGCCACCATCAGTGATGTAAACCTGGAGTCAGAATTGAGATGTGCCCTGACTGAATACACCCCACAGTTCTCGAGAATAGTTCAGTCTGTTAAATACCAGTATTCTCATTGACTTGATCAAAATTGATCAATGACTAGCACTAGGCCTATAACTGTGCACTTTTGTATGAATAAAGTTGATTAAACTGTTTTTTGGACTGCTTTTGTATTACTGCTNAAAATCCATATTTTTGAATAGCTAACAGGCTATATTTTCACCATTGTTATCAATAAAAAAATTCATGCGGCCCGCACACATATGGATTTCTGATCATGTGGCCCACTATTGAAAAAACNTGGACGCCTCTGCTCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie13a | ATF4 | 278 | 287 | - | 17.50 | ATGATGCAAT |
| Charlie13a | CEBPG | 278 | 287 | - | 17.31 | ATGATGCAAT |
| Charlie13a | SPIB | 343 | 355 | - | 16.54 | TCACTTCCTCCAT |
| Charlie13a | Prdm4 | 632 | 642 | + | 16.16 | ACTTGAAACTG |
| Charlie13a | atfs-1 | 278 | 287 | - | 15.96 | ATGATGCAAT |
| Charlie13a | Znf423 | 45 | 59 | - | 15.75 | CGCACCCTGGGGTCA |
| Charlie13a | DUXA | 1269 | 1281 | - | 15.57 | TTGATCAAGTCAA |
| Charlie13a | DUXA | 1269 | 1281 | + | 15.55 | TTGACTTGATCAA |
| Charlie13a | Prdm4 | 955 | 965 | - | 15.50 | TCTTGAAACTG |
| Charlie13a | SPIB | 961 | 973 | - | 15.49 | TCAGTTCCTCTTG |
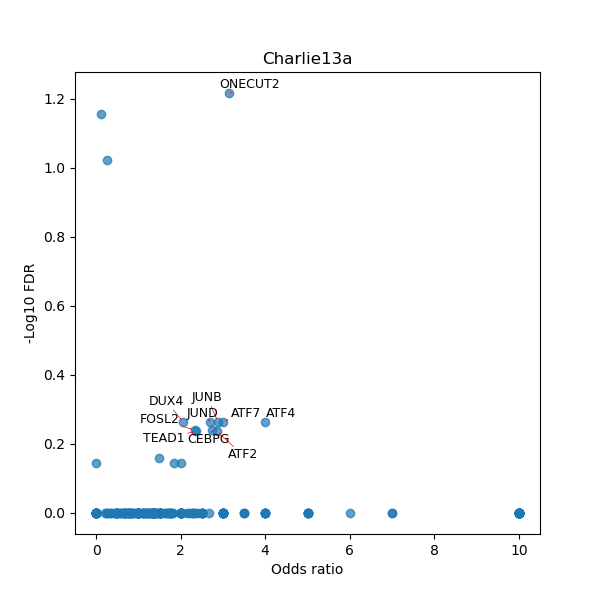
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.