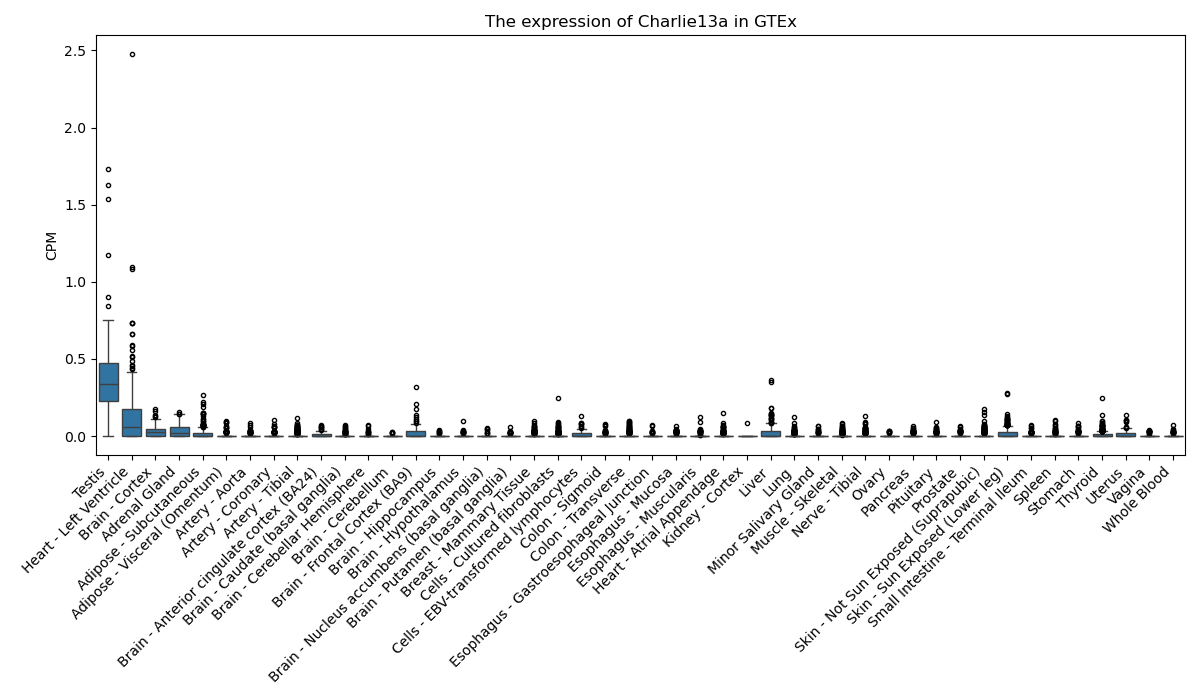
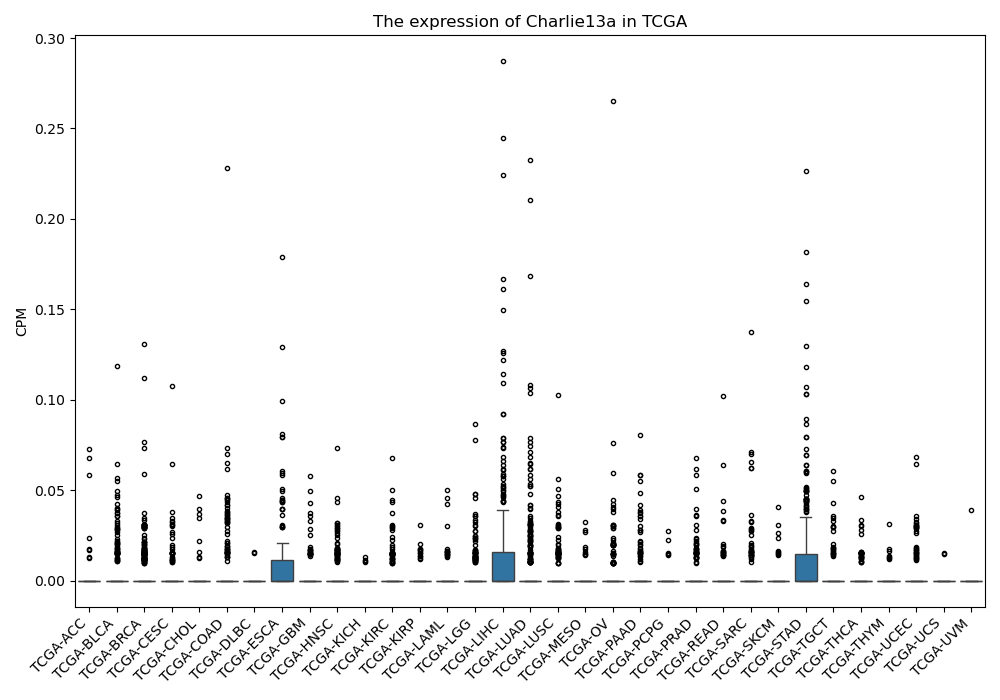
Charlie13a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000086 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Mammalia |
| Length | 1514 |
| Kimura value | 32.78 |
| Tau index | 0.9930 |
| Description | hAT-Charlie DNA transposon, Charlie13a subfamily |
| Comment | - |
| Sequence |
CAGGGACATCCAATATGCAGCCCGCGGGCCAAATGCGGCCCGCCTGACCCCAGGGTGCGGCCCGCCTGAGATTTTTAGCAAAAATGTTTTTACCTAATTAGCTAGCACACATGAACTAGTAGGGCCTAACCACCTGCCAGTAAGGAATTTGAGAAGTTTGGTCTAAACATGGAAAAGCTGAGTTCACTTGCCACAGATGGGGCACCTGCAATGAAAGGGAAGAAAGGTTTCAACAGTTTATTCAAGTCAAAAGAGAAGGTTGGTGTGCCCAGCTTCCATTGCATCATTCATCAGGAGAGTTTGTGCTGTGAACTTTGCAAGTCAGGTTCATTGCATGATGTGATGGAGGAAGTGATCAAGATTGTGAATTTTCTTCGTGCTCGTGCCCTTAACCATCGCCAGTTTATGGGATATTTGGAAGAAGTTGAAGCAGAGTATGGAGATTTAGTTTATTTTAATGCAGTCAGATGGCTAGGTCGTGGAAACTTGTTGAAGAGATTTACTGAGCTGCTTCCTCAGATCAAGGACTTTTTGGAATTAAAGGGCTCTGAGAGACCCAACTTGTCGGACCCCCAGTGGCTCACAAGATTGTACTTTCTTACTGATGTGACAAGACATCTGAACACACTGAACTTGAAACTGCAGGGAAAAAACAAGAGCATTGCTGATTTGTTCAAGGAAGTGCAGGTATTCAGACTCAAACTGGACATGTGGATTGAGCAGATGGCAACTGGAGACTGCACTCATTTCCCATTGCTGAACTGCCCTGAGCTCAATGGAATCACAGATTTTGAAGAACTGCAGAGCTATTTGGTGGAAATGAAGTCACAATTTCAGAAGAGATTCACAGATTTTGATGAGTATGAATCATGCTTTAAGTTTCTGCGGCTGCCTTTGGAGTGCAAACCTCAGGATGTGTCTGAATTATCAAGCATCTGCCCTATCAGTGTGGCTCAGTTTCAAGAGGAACTGATTGAGCTGAAAGCGAACTATGCCCACCAAGAAGACACAGTATCCCTGGAATTTTGGAGATGAGTATATGAATCCCAGAACTATCCTGAAATTTCTGGCTATGTGGCCAAACTCTGGGCAATGTTTGGGTCCACCTGGCTCTGTGAAAGTACATTCTCTTTCATGAAACTGCTGAAATCGAAGCTGAGAGCCACCATCAGTGATGTAAACCTGGAGTCAGAATTGAGATGTGCCCTGACTGAATACACCCCACAGTTCTCGAGAATAGTTCAGTCTGTTAAATACCAGTATTCTCATTGACTTGATCAAAATTGATCAATGACTAGCACTAGGCCTATAACTGTGCACTTTTGTATGAATAAAGTTGATTAAACTGTTTTTTGGACTGCTTTTGTATTACTGCTNAAAATCCATATTTTTGAATAGCTAACAGGCTATATTTTCACCATTGTTATCAATAAAAAAATTCATGCGGCCCGCACACATATGGATTTCTGATCATGTGGCCCACTATTGAAAAAACNTGGACGCCTCTGCTCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie13a | NR1I2 | 620 | 634 | + | 15.43 | TGAACACACTGAACT |
| Charlie13a | CUP2 | 76 | 86 | + | 15.41 | TAGCAAAAATG |
| Charlie13a | TCP16 | 1099 | 1106 | - | 15.32 | GTGGACCC |
| Charlie13a | MYB80 | 89 | 100 | + | 15.26 | TTTACCTAATTA |
| Charlie13a | MYB49 | 90 | 100 | + | 15.18 | TTACCTAATTA |
| Charlie13a | FERD3L | 190 | 203 | - | 15.00 | GCCCCATCTGTGGC |
| Charlie13a | TCF4 | 202 | 209 | + | 14.77 | GCACCTGC |
| Charlie13a | ASCL1 | 130 | 138 | + | 14.74 | CCACCTGCC |
| Charlie13a | Neurod2 | 193 | 200 | + | 14.69 | ACAGATGG |
| Charlie13a | FOXH1 | 710 | 717 | - | 14.67 | AATCCACA |
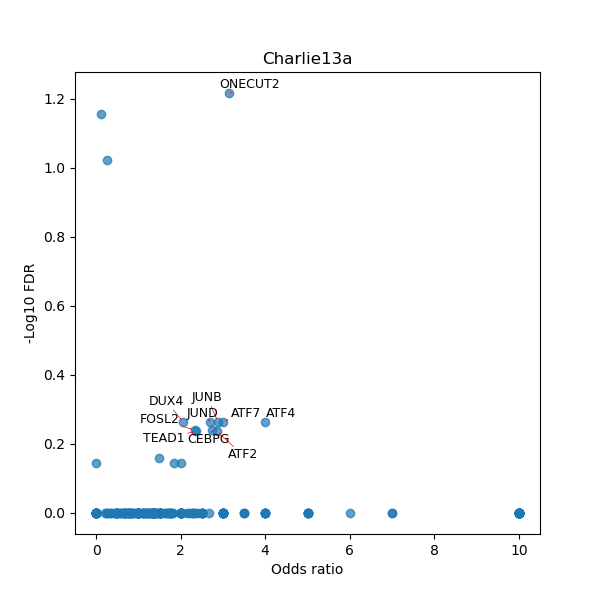
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.