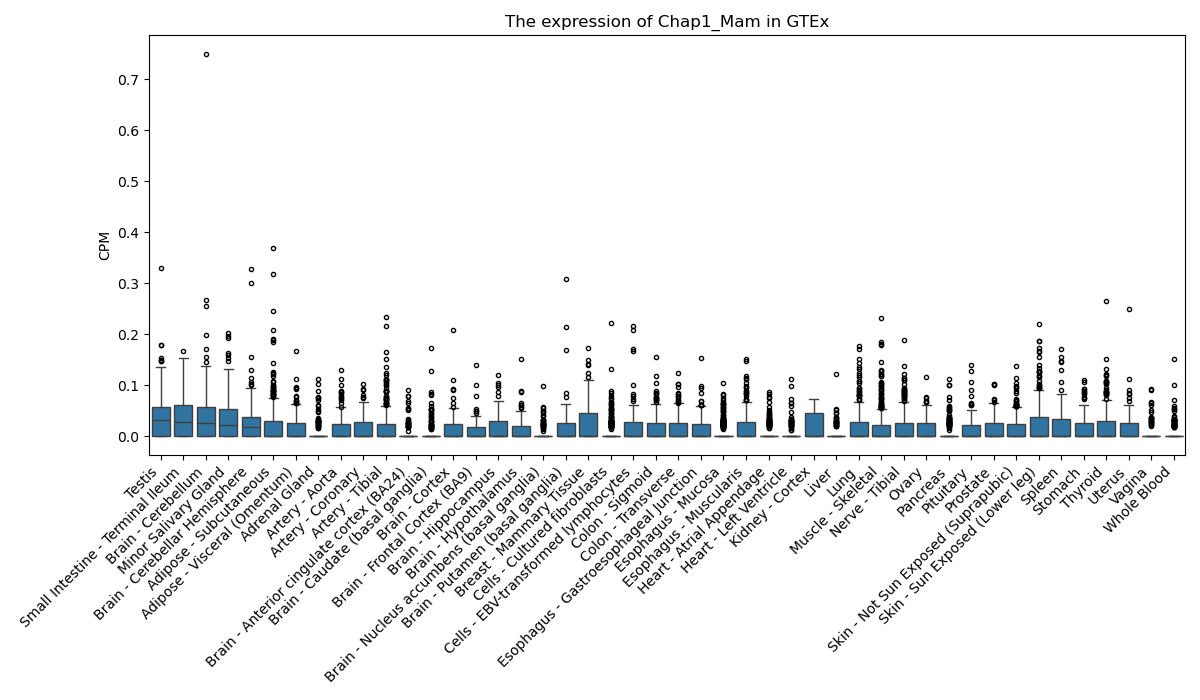
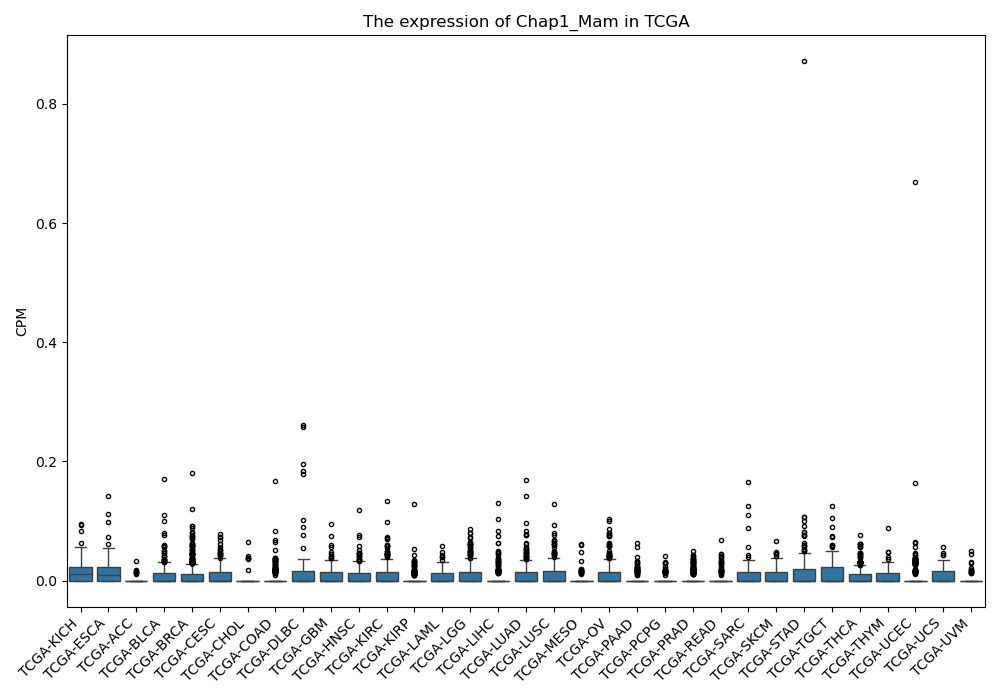
Chap1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000080 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 342 |
| Kimura value | 29.70 |
| Tau index | 0.9332 |
| Description | hAT-Charlie DNA transposon, Chap1_Mam subfamily |
| Comment | Chap1_Mam TSD is "NNCTAGNN". |
| Sequence |
CAGCGGTTCTCAACCGCGGTGTCACGACACACTGGTGTGTCGCGATCAGTTGTGAGGTGTGTCGCAAGTAATTTGTTACTAATAATAGTTAAAGTACCGAAGTTTGCGAATGATTTACTTTTTTTAAATAAATTAGAGCGGATTCAAGGTTATCCCTATAATAACGTCACTCTCACTCACTTGCATTCCGATATGCTTTCTTGAGTGGGAGAGAAAGGGGTGGAGTTACAGCTAGCGCGCCGGGAATTGCGCGTAACCCCGGGTATGCCCATGGGAGTGTGTCGCGCATGTGAACGTCATCCGTCACGTGTGTCGCGGCGGAAAAAAGGTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Chap1_Mam | knrl | 130 | 140 | + | 13.97 | AAATTAGAGCG |
| Chap1_Mam | MLXIPL | 303 | 310 | + | 13.82 | GTCACGTG |
| Chap1_Mam | DOF4.7 | 115 | 123 | + | 13.72 | TTACTTTTT |
| Chap1_Mam | MEF2D | 78 | 89 | + | 13.71 | ACTAATAATAGT |
| Chap1_Mam | Fer2 | 302 | 311 | - | 13.69 | ACACGTGACG |
| Chap1_Mam | Ebf2 | 268 | 276 | + | 13.62 | CCCATGGGA |
| Chap1_Mam | ZAT10 | 168 | 177 | + | 13.55 | CACTCTCACT |
| Chap1_Mam | Blimp-1 | 168 | 177 | + | 13.52 | CACTCTCACT |
| Chap1_Mam | TYE7 | 304 | 310 | - | 13.51 | CACGTGA |
| Chap1_Mam | D1 | 124 | 131 | + | 13.38 | TTAAATAA |
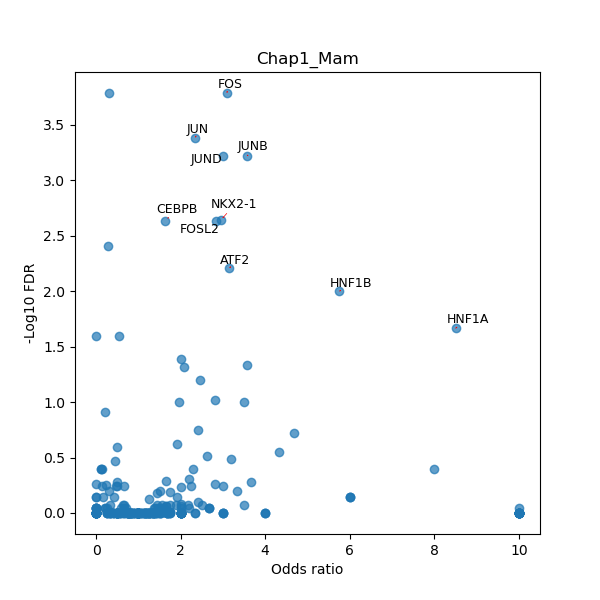
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.