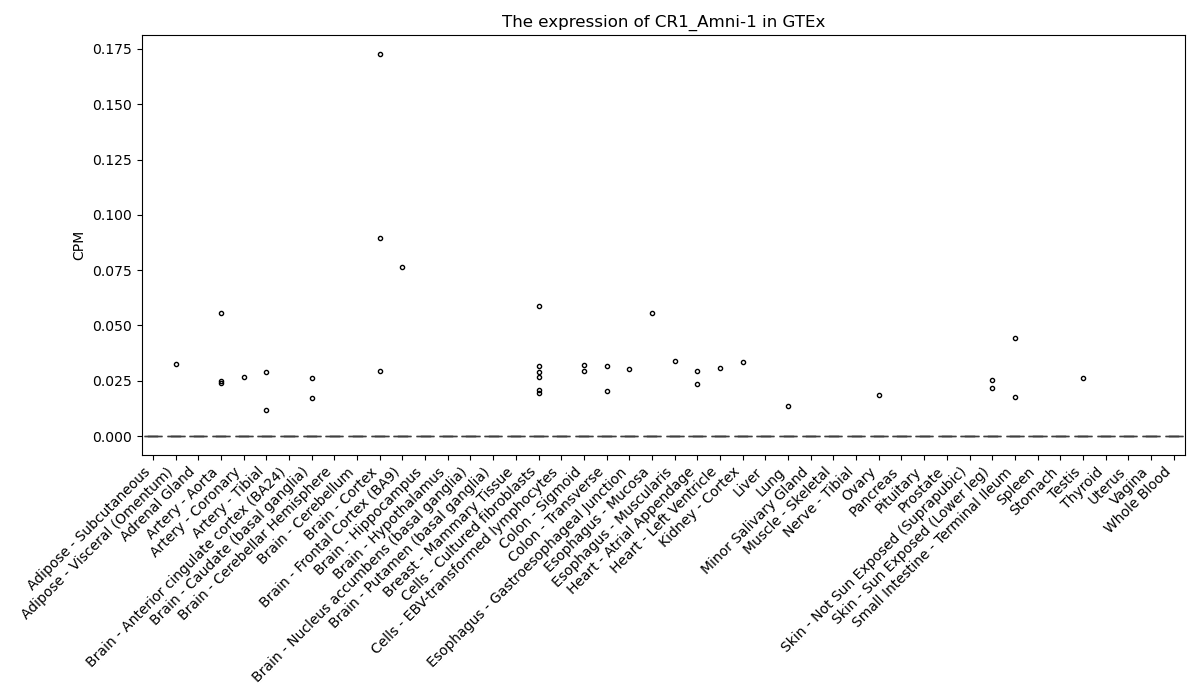
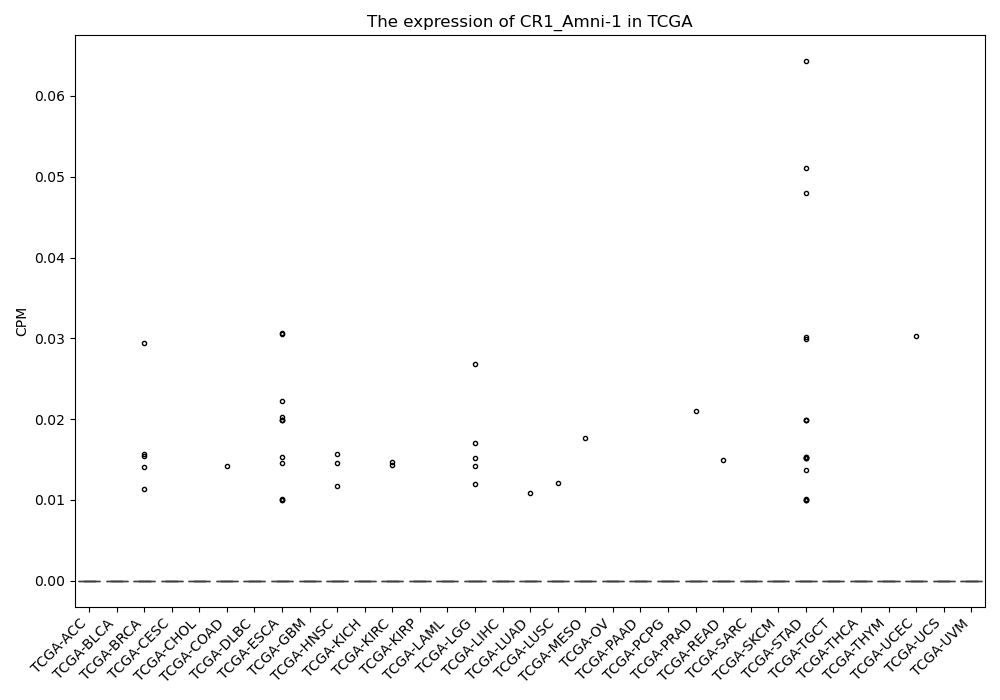
CR1_Amni-1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0004193 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 1319 |
| Kimura value | 33.73 |
| Tau index | 0.0000 |
| Description | CR1 LINE element |
| Comment | Possibly ancestral CR1 element to CR1 elements in Sauropsida and mammals. Closest matches to CR1-1(B)_CPB, which may be common to all turtles, CR1-15_AMi and CR1-14_Ami (common to all Crocodylia), the monotreme PlatCR1 and mammalian-wide CR1_Mam. Position 879-1172 was originally entered as X6B_LINE. The final 100 bp (pos 1223-1320) are very conserved amongst the descendant CR1 elements. For an orthologous copy in all amniotes see for example hg38 chr16:72,993,631-72,994,220. |
| Sequence |
GGTGGACATAATTTACTTGGATTTTCAAANAGCTTTTGATAAGGTCTCTCACAAAAGATTATTAAGGAAAATAAATAACCANAGGGTAAGGGGCAAAGTNCTGTCGTGGATTAAAAACTGGTTAAACGATAGGAAACAAAGANTCGCAGTGAATAGCCGTTCTCAGAGTGGAGAGAGGTTAATAGCGGGNTACCCCAGGGGTCTGTATTGGGACCGGTGTTATTTAATATATTTATTAATGATTTGGAGGAGGAAGTGGACAGTAAGTTGGCGAAGTTTGCTGATGACACTAAGTTGTTTTGGTTAGTAAAAACCACGGAGGATTGCGAGAAACTCCAGATGGATTTAAATACACTGGGGGATCGGGCAGCACGATGGCAGATGAAATTCAACTTGGATAAGTGTAAGGTAATGCACATTGGCAAAAATAGTCTAAACTTCTCATGTACGCCGATGGGCTCTGAACTTGCGGGATCCTCTGAGGAAAAAGACTTAGGAGTCATCGCGGATAGCTCAATGAAGACGTCTGCCCAGTGCGCAGCGGCAGTTAAAAAAGCAAATAGGATGCTTGGGTGCATTAAAAAGGGAATGGAAAATAATACAGAAAATATTATAATGACATTATATCGAACAATGGTACGCCCTCATCTGGAGTACTGTGTCCAGTTTTGGTCGCCTCACTTTAAGAAAGATATAGCGGAGATTGAAAAGGTCCAGAGAAGGGCAACGAAGATGGTTAGAGGTATGGCGGGACTTACGTATGATGAGAGACTGCAAAAACTAGGATTATTTAGCCTGGAAAGGAGAAGAGTTAGAGGGGACTTGATTGAGGTATTCAAAATTATGAAGGGTATAGTGAAAACTGATCAGTCCCTCCTTTTTACCCTGTCCCATAATACGAGAACAAGGGGTCACTCGATGAAATTAANGGNCGGTAAATTTAGAACAAATAAAAGGAAATACTTCTTCACGCAGCGTGTAGTCAGCCTGTGGAATTCACTGCCGCAGGAGGTCATCGAGTCAAATACTGTGGCTGGATTTTNAAAAGGGCTGGATAATTTTATGACCANTAATAACATTTGTAGTTATGCANGCTAAGATAAGGGTAATCAAATCTCATGCTTCAGGGCGTAAGCTGATCGCCTGCAGGGGTCAGGAAGGAATTTTTCCCCCACGTACAGCATTGCACAATTGGCTAGGTGCATTACGGGGTNTTTTCGCCTTCCTCTGAAGCATCAGGTATTGGCCACTGCCGGAGGCAGGATACCGGACTNGATGGACCAATGGTCTGATCCGGTATGGCAAATCCTATGTTCCTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Amni-1 | HOXC9 | 1059 | 1067 | - | 13.90 | GTCATAAAA |
| CR1_Amni-1 | KUA1 | 395 | 402 | - | 13.89 | CTTATCCA |
| CR1_Amni-1 | TFAP2B | 1252 | 1260 | - | 13.85 | GCCTCCGGC |
| CR1_Amni-1 | opa | 1003 | 1014 | - | 13.85 | GACCTCCTGCGG |
| CR1_Amni-1 | Sox17 | 628 | 637 | + | 13.84 | CGAACAATGG |
| CR1_Amni-1 | DOF3.4 | 551 | 567 | - | 13.83 | CATCCTATTTGCTTTTT |
| CR1_Amni-1 | HOXB9 | 1059 | 1067 | - | 13.79 | GTCATAAAA |
| CR1_Amni-1 | FOXD3 | 64 | 77 | + | 13.79 | AAGGAAAATAAATA |
| CR1_Amni-1 | ZFP57 | 1001 | 1007 | + | 13.74 | TGCCGCA |
| CR1_Amni-1 | ZNF282 | 886 | 900 | + | 13.73 | CTGTCCCATAATACG |
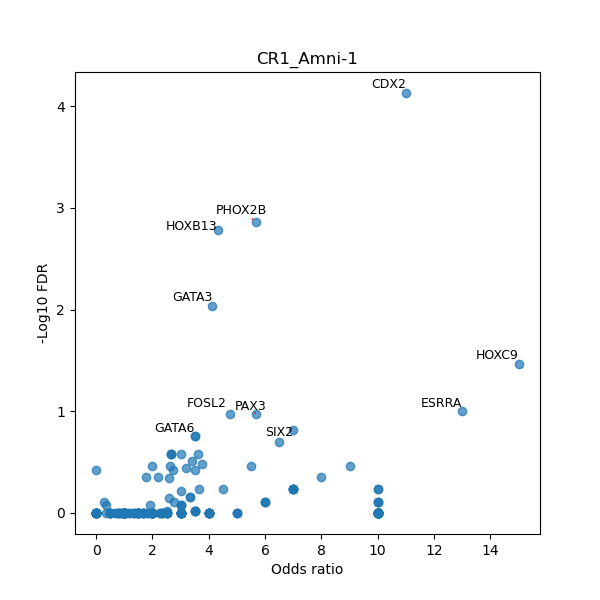
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.