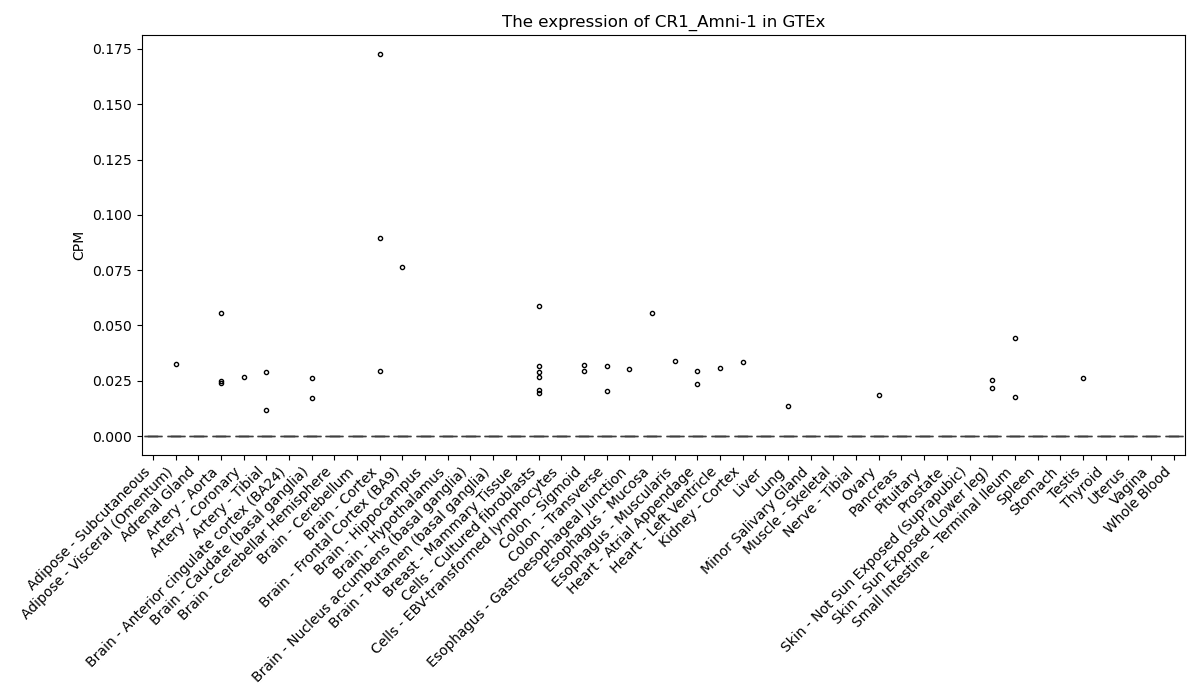
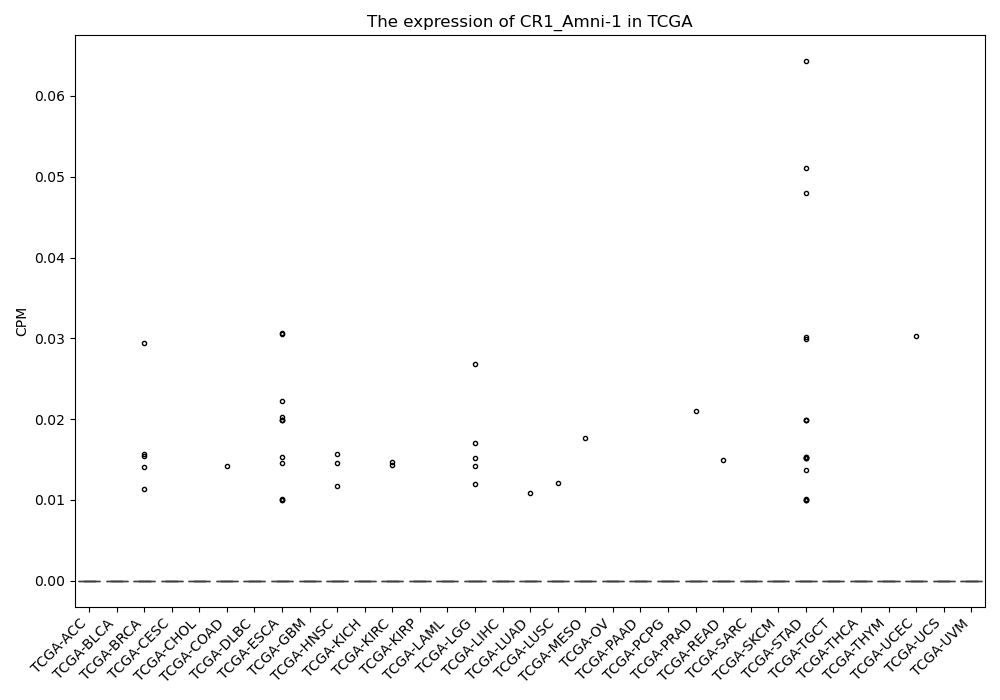
CR1_Amni-1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0004193 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 1319 |
| Kimura value | 33.73 |
| Tau index | 0.0000 |
| Description | CR1 LINE element |
| Comment | Possibly ancestral CR1 element to CR1 elements in Sauropsida and mammals. Closest matches to CR1-1(B)_CPB, which may be common to all turtles, CR1-15_AMi and CR1-14_Ami (common to all Crocodylia), the monotreme PlatCR1 and mammalian-wide CR1_Mam. Position 879-1172 was originally entered as X6B_LINE. The final 100 bp (pos 1223-1320) are very conserved amongst the descendant CR1 elements. For an orthologous copy in all amniotes see for example hg38 chr16:72,993,631-72,994,220. |
| Sequence |
GGTGGACATAATTTACTTGGATTTTCAAANAGCTTTTGATAAGGTCTCTCACAAAAGATTATTAAGGAAAATAAATAACCANAGGGTAAGGGGCAAAGTNCTGTCGTGGATTAAAAACTGGTTAAACGATAGGAAACAAAGANTCGCAGTGAATAGCCGTTCTCAGAGTGGAGAGAGGTTAATAGCGGGNTACCCCAGGGGTCTGTATTGGGACCGGTGTTATTTAATATATTTATTAATGATTTGGAGGAGGAAGTGGACAGTAAGTTGGCGAAGTTTGCTGATGACACTAAGTTGTTTTGGTTAGTAAAAACCACGGAGGATTGCGAGAAACTCCAGATGGATTTAAATACACTGGGGGATCGGGCAGCACGATGGCAGATGAAATTCAACTTGGATAAGTGTAAGGTAATGCACATTGGCAAAAATAGTCTAAACTTCTCATGTACGCCGATGGGCTCTGAACTTGCGGGATCCTCTGAGGAAAAAGACTTAGGAGTCATCGCGGATAGCTCAATGAAGACGTCTGCCCAGTGCGCAGCGGCAGTTAAAAAAGCAAATAGGATGCTTGGGTGCATTAAAAAGGGAATGGAAAATAATACAGAAAATATTATAATGACATTATATCGAACAATGGTACGCCCTCATCTGGAGTACTGTGTCCAGTTTTGGTCGCCTCACTTTAAGAAAGATATAGCGGAGATTGAAAAGGTCCAGAGAAGGGCAACGAAGATGGTTAGAGGTATGGCGGGACTTACGTATGATGAGAGACTGCAAAAACTAGGATTATTTAGCCTGGAAAGGAGAAGAGTTAGAGGGGACTTGATTGAGGTATTCAAAATTATGAAGGGTATAGTGAAAACTGATCAGTCCCTCCTTTTTACCCTGTCCCATAATACGAGAACAAGGGGTCACTCGATGAAATTAANGGNCGGTAAATTTAGAACAAATAAAAGGAAATACTTCTTCACGCAGCGTGTAGTCAGCCTGTGGAATTCACTGCCGCAGGAGGTCATCGAGTCAAATACTGTGGCTGGATTTTNAAAAGGGCTGGATAATTTTATGACCANTAATAACATTTGTAGTTATGCANGCTAAGATAAGGGTAATCAAATCTCATGCTTCAGGGCGTAAGCTGATCGCCTGCAGGGGTCAGGAAGGAATTTTTCCCCCACGTACAGCATTGCACAATTGGCTAGGTGCATTACGGGGTNTTTTCGCCTTCCTCTGAAGCATCAGGTATTGGCCACTGCCGGAGGCAGGATACCGGACTNGATGGACCAATGGTCTGATCCGGTATGGCAAATCCTATGTTCCTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Amni-1 | GATA4 | 1098 | 1105 | - | 14.20 | CCTTATCT |
| CR1_Amni-1 | Nfat5 | 589 | 596 | + | 14.18 | ATGGAAAA |
| CR1_Amni-1 | ZFP14 | 249 | 263 | + | 14.17 | GGAGGAAGTGGACAG |
| CR1_Amni-1 | ELF3 | 250 | 258 | - | 14.16 | CACTTCCTC |
| CR1_Amni-1 | EHF | 250 | 258 | - | 14.15 | CACTTCCTC |
| CR1_Amni-1 | EcR | 1009 | 1016 | + | 14.10 | GAGGTCAT |
| CR1_Amni-1 | Nfatc2 | 588 | 595 | + | 14.03 | AATGGAAA |
| CR1_Amni-1 | usp | 1149 | 1157 | + | 14.00 | GGGGTCAGG |
| CR1_Amni-1 | CDX4 | 1059 | 1067 | - | 14.00 | GTCATAAAA |
| CR1_Amni-1 | usp | 908 | 916 | + | 13.90 | GGGGTCACT |
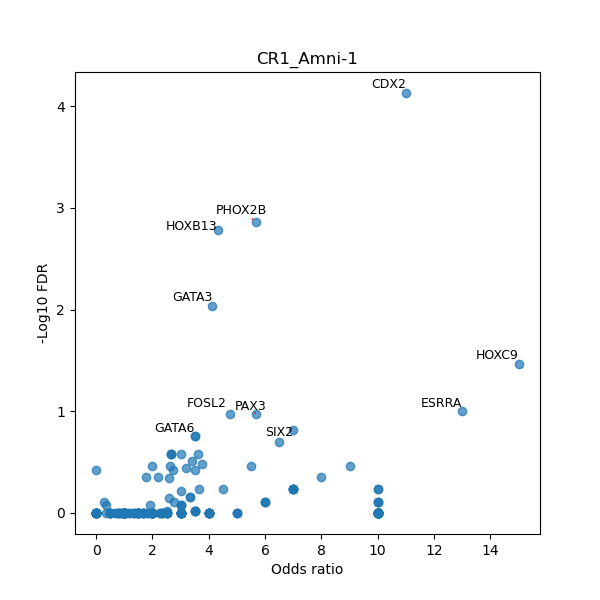
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.