CR1-L3A_Croc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001326 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 4288 |
| Kimura value | 35.65 |
| Tau index | 0.0000 |
| Description | CR1 (Chicken Repeat 1) retrotransposon, CR1-L3A_Croc subfamily |
| Comment | - |
| Sequence |
CTGTGACCCTTGGGTCTNTGATCAGCCCTGGTGTTTGAAGTCTCTGAGTAGTTAATCCCTCCCTGATTAGCGAGGGGGTGGAGCTGAGCAGCGAGGCTTAGTAGTAAAGTCAGTTGCCAGGCGAACTCNTGAGCAGCGGAAAACGAGAGCGTTTTGGAGGGAGTTTGGAGGGGAGTGTGNGAAGCTTTCTTTCTAGGTACAGTTCTACGCTCAATACCGTGATGGCCAGCGATGGANCAGTGGTTGTGACCTGCAACGGATGCGCCACGTTTTCTTTCCTGCCTGAAGCCAGAAGGGACTTCCTGTGTATGAAGTGCAAGTTGGTGGCTGTGCTGGAAGAAAAGATTCGTGGTTTGGAAGGACAAGTACTGACGCTGCGGAGCATCAGAGAGGCTGAGGAGTTCTTGGACAACCAGATTCGGGAAGCATCACCACCCCAGATTCAAGAAAGAAAGGAACTGGCCACCAAGGAACCAGAAAGAGAGGAAACCAGAGAGGAGGCTTGGCAGTTCGTAACCACCAGAGGCAAGAGGACGAGGCGGCATTCTACACGGCTGGAGGCAGANGAGGATGGGCAGGCTGTGACCACCAGAGAGAAGAGGACCAGGAGGAATTCCACACAGCTAGAAGTATCCAATCGTTACCAGGTTCTCAGCGTGGAAACTGCAGAAAATATCTCTGAAGATCCAGCTGGTCGAATGGAACGGAGAGCTGTACGGGACATACCTGTGGATGGCTGCTCAGCCCAACCCGCAAGACGATCAAGCGTGCCCACAAAGAGATCTCCAACCATCCAAGGAAGACAGACGATCCTTGTCGGGGATTCTATACTAAGAAGAATGGAAAGAACGTTCTGCAAGGGACAGGCGGACAACAGGACGGTGTGCTGTCTTCCCGGAGCCAAGACACGAGATGCGACTCTAAGACTGGATAGGCTTCTGAAGTCGGCGGGCAAGGATCCACTGGTGATGGTGCATATCGGCACTAATGACACTGCTTCGCGNGATACCTCACAGATTATAGACGACTTCAGGGAACTCGGAAGGGTGCTGAAGGAGAAGAATGTCCAAGTGATCTTCTCGGAGATCCTTCCTGTCCCACGAGCGAAGGAAGACAGAAGGCAGAAGATTCTGGAAGTGAACCGCTGGCTAGGTAAGTGGTGTAAGGTAGAGGGTTTTGGTTTTGTGGAACACTGGTCCACCTTCTATGGGGAGAAGGGGCTGTACAGTTTGGATGGCCTCCATCTCAGTAGAAGGGGAACCAGTCTTCTAGGGGACAGGCTGGCTAGACTAGTCAGGAGGGCGTTAAACTAACAACAAAAGGAGGGAGTGAAAGAGGGAAACAAATGAGCACTCATTTAGCACAAAATCAAGATGTCGAGAACAAAATTAATCAAGGCACCAAGGGACGTGAAGAGAAGAAATTCTTTAATTGCCTATACACCAATGCTAGGAGCCTGGGTAATAAACAAGAGGAATTGGAATTGCTCATTTATGAGCATAAATTCGACCTAGTTGGTATTACTGAAACCTGGTGGGAAGATTCGCATGACTGGAATGTTAAAATCAATGGTTATAACCTATTTAGGAAGGATCGAGTGGGCAAAAGGGGAGGGGGAGTGGCACTCTATGTCAAAAATGGCATTACCTGTTTCCGAGTCACTGACAACTCGGAAGCAAATGATCTTGAATGCTTATGGATCAATGTTCTAACAGATAAAGCACAAGATGGGGTACTAGTTGGTGTCTGCTACAGACCACCAAATCACACTAGAGAACAGGATGACCGGCTCCTTAAGCACCTATCTATAATGTGTAGGGGAAAAAGCTGCGTTATCACGGGGGACTTCAATCTGAGTGACATATGCTGGAGGTCTCATGCTGCCAGTACTAAAACGTCCTCGGAATTTCTAAAAATTATAGATGACAATTTCCTAACTCAAAAAGTGTTGCATCCAACACGGGGGAATTCTATATTAGACCTCATCTTGACAGATAAAGAGGAATTGATCACAGAACTAAAAATTAGTGGTTGCTTAGGTGCAAGTGATCATGACTTGATTACATTTGTTATGTGCAAACAGAATAAAGTCCAAACCAGTAATATATATACTTGGTGCTTTAAAAGGGCCAGTTTCACAAAGCTGAAAACAATTATGAGCCAAATCAGCTGGGAGAAAGAATTTAAACAGAAAAATGTGAATGATAATTGGGAACTGTTTAAGAACATTTTACTAGATGCCCAAAAAGCCACAATCCCACAATCGAGAAAGAAGGCTACGCTGGTTAAAAAACCAGCCTGGCTTAGAGGGGAAGTGAAAGCAGCTATAAAAAATAAAAAAGCAATATATAACAAATGGAAAAAAGGGGAAGTTGATAGCAATGAATATAAATTAGAAGCTAGGAATTGTAGAAAATTGATAAGGGAAGCAAAAGGACACAAGGAGAAATCTATGGCCAGCAGAGTTAAGGACAATAAGAAGGAGTTTTTNAAGTATATTAGGAACAAAAGGAATCCTAACAATGGTATTGGTCCATTACTAGATGGAAATGGTAGAATTGTCAATAATAATGCAGAAAAGGCAGAAGTGTTCAATAAATATTTCTGTTCTGTATTTGGGAAAAAGCCAGATGATGTANTCGTATCACATGATGATGATGAAATACTTTCCATTCCAACAGTAACTAAGGAGGATGTTAAACAGCAGCTACTAAAGTTAGACATTTTTAAATCAGCAGGTCCGGATAACTTGCATCCAAGAGTTTTAAAAGAGCTGGCCGAGGAGCTCTCTGGACCGTTAATGTTGATTTTCAATAAGTCTTGGAACACTGGGGAAGTTCCAGAGGACTGGAAGAAAGCTAATGTTGTGCCAATATTTAAAAAGGGTAAACGGGATGACCCGGGTAATTATAGGCCTGTCAGCCTGACATCGATCCCGGGCAAAATAATGGAACGGCTGATACGGGACTCGATTAATAAAGAATTAAAGGAGGGTAATATAATTAATGCCAATCAACATGGGTTTATGGAAAATAGATCTTGTCAAACTAACTTGATATCTTTTGATGAGATTACAAGTTTGGTTGATAAAGGTAATAGCGTTGATGTAATATACTTAGACTTCTGTAAGGCATTTGACTTGGTACCGCACGACATTTTGATTAAGAAACTAGAACGATACAAAATCAACATGGCACACATTAAATGGATTAAAAACTGGCTAACTGATAGGTCTCAAAATGTAATTGTAAATGGGGAATCATCATCGAGCGGGCGTGTTTCTAGTGGGGTCCCGCAGGGATCGGTTCTTGGCCCTACGCTATTTAACATTTTTATCAATGACCTGGAAGAAAACATAAAATCATTACTGATAAAGTTTGCAGATGACACAAAGATTGGGGGAGTGGTAAATAATGAAGAGGACAGGTCACTGATACAGAGCGATCTGGATCGCTTGGTAAGCTGGGCGCAAGCAAACAATATGCGTTTCAATACGGCCAAATGTAAAGTCATACATCTAGGAACAAAGAATGCAGGCCATACTTACAGGATGGGGGACTCTATCCTGGGAAGCAGCGACTCTGAAAAGGACTTGGGGGTCACGGTGGATAATCAGCTGAACATGAGCTCCCAGTGCGACGCTGTGGCCAAAAGGGCTAATGCGATCCTTGGATGTATAAACAGGGGAATNTCGAGTAGGAGTAGGGAGGTTATATTACCTCTGTATTTGGCACTGGTGCGACCGCTACTGGAATACTGTGTCCAGTTCTGGTGTCCACANTTCAAGAAGGATGTTGANAAATTGGAGAGGGTTCAGAGAAGAGCCACGAGAATGATTAAAGGACTGGAAAACATGCCTTATAGTGAGAGACTCAAGGAGCTCAATCTATTTAGCTTATCAAAGAGAAGGTTAAGGGGTGACTTGATCGCAGTCTATAAGTACCTACACGGGGAACAGAAATTTGATAATAGAGGGCTCTTCAGTCTAGCAGACAAAGGTATAACAAGATCCAATGGCTGGAAGTTGAAGCTAGACAAATTCAGACTAGAAATAAGGNGCAANTTTTTAACAGTGAGGGTAATTAACCACTGGAACAACTTACCAAGGGNTGTGGTGGATTCTCCATCACTGGAAATTTTTAAATCAAGATTGGATGTTTTTCTAAAAGATATGCTCTAGTTCAAACAGGAATTAATTCGGGGAAGTCCTATGGCCTGCGTTAGCAGGAGGTCGGACTAGATGATCTACAGNGGTCCCTTCCGGCCTTANAATCTATGAATCTATGAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-L3A_Croc | HSFA1E | 1123 | 1136 | + | 19.51 | AGAAGATTCTGGAA |
| CR1-L3A_Croc | Sox1 | 2519 | 2533 | - | 18.91 | ACCAATACCATTGTT |
| CR1-L3A_Croc | Sox21a | 2519 | 2533 | - | 18.79 | ACCAATACCATTGTT |
| CR1-L3A_Croc | HSFA6A | 1124 | 1136 | + | 18.69 | GAAGATTCTGGAA |
| CR1-L3A_Croc | Hmx2 | 1423 | 1437 | - | 17.90 | AGGCAATTAAAGAAT |
| CR1-L3A_Croc | HSF1 | 1124 | 1136 | - | 17.84 | TTCCAGAATCTTC |
| CR1-L3A_Croc | SPIB | 2306 | 2318 | - | 17.78 | TCACTTCCCCTCT |
| CR1-L3A_Croc | HSF2 | 1124 | 1136 | - | 17.38 | TTCCAGAATCTTC |
| CR1-L3A_Croc | SoxN | 2521 | 2531 | - | 17.36 | CAATACCATTG |
| CR1-L3A_Croc | DOF3.6 | 2326 | 2346 | - | 17.15 | ATTGCTTTTTTATTTTTTATA |
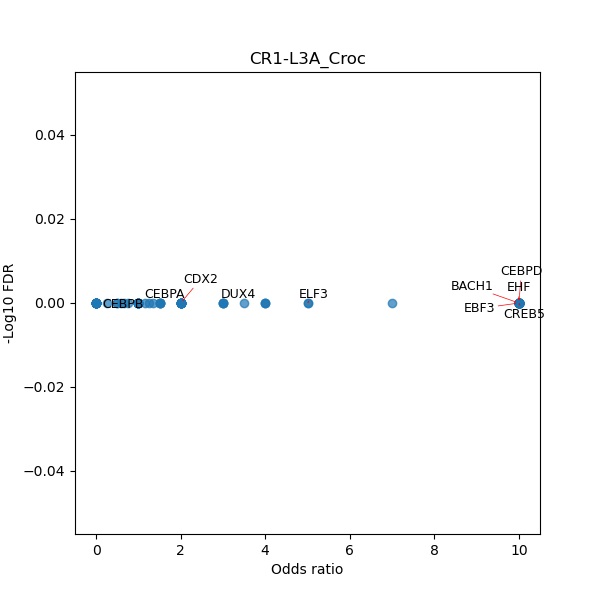
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.