CR1-3_Croc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001325 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 3605 |
| Kimura value | 33.02 |
| Tau index | 0.0000 |
| Description | CR1 (Chicken Repeat 1) retrotransposon, CR1-3_Croc subfamily |
| Comment | - |
| Sequence |
GGGATCCATCTGACGAGAACGGGGAAGAGCATCTTTGGGCGTCGTCTGGCCAACCTAGTAAGGAGGGCTTTAAACTAGGTTCCACGGGGATGGTGATCAAAACCCTCCAGCAACAGAGCACACAGTCCAGAGGGGAAAGGACTCGAAGAAGCAACTCAACTCTGGGAAAGGGCCAGAGAGCTACAGGACTGCAACAAGTGGCCAAAAGGAAGCGAGTAGGTCAAAATGCAAAATACGTCCGATGCATGTATACAAATGCGAGAAGTATGGGGAATAAACAAGATGAGCTGGAAGCTGCAGCCTGTGAAAAGAACTATGATCTGATTGGCATCACAGAGACCTGGTGGGACAGCTCTTATGACTGGAATGTCAACATCGAGGGTTACACTCTGTTTCGGAAGGACAGGGTTGGGAGAAAAGGTGGTGGTGTTGCCTTGTATGTCAAAAACACATACACTTGTTCCCTGGTTCAGGAGGAAGAAGACAGCAAAGCAGAATGCATTTGGGTGAAGATGAAAGGTGAAAGATGCAGCACCGATATAATGGTGGGGATTTATTATAGGCCACCAAATCAGGAAGAGGAGGTGGATGAGGCACTCTTCAAGCAGGTGACAAATCTATCCAAAAGCTATGGGATGGTACTGATGGGAGACTTCAATTTTCCGGACATCTGCTGGAAGAACAATGCAGCCAAACATAAAATGTCAAGCCGGTTCCTAACTTGTGTGGAGGACAACTTTCTGTTCCAGAAAGTGGAGGATACAACTAGGGGATCATCTATCTTGGATCTTGTCCTGACCAACAGGGAAGAACTGGTGGAGGATGTGAAGGTGATGGGTAACTTGGGAGGAAGCGATCACGATATGATAGAATTCCAGATCCTGAGACGGGGAAAGCATGAGGTCAGCAAAATTAGGACGCTAGATTTCAAAAAGGCGGATTTCATCCGACTCAAGGAGTTAATAGGCATGGTGCCATGGGAGGACAGACTGAAGGATAAAGGCGCCCAGGAGGGCTGGGAGCTTCTAAAGGGCACAGTATTGGAAGCCCAACAAGAAACTGTCCCGACACGATGGAAGCACAAGAAGAATGGCAAGAGACCAATGTGGCTGCACAAGGAGCTTCTAAAATGCCTCAAATGCAAAAGGAAAGCATACAGGCAATGGAAGAATGGGCAGGTCACCAAGGAAGCGTACCAGGAAATAGCAAGAACCTGCAGGGACAAAATCAGGAAGGCAAAGATAAAGAACGAGCTGCACCTGGCGAAGGAGGTTAAGGACAACAAGAAGAGGTTCTATAAGTATGTTGGCCAGAAAAGAAGGACCAAGGAAGCTGTGGGTCCNCTGTTAACAAGCCAGGGGGAACTCCTAACNGAAGATGCAAAGAAGGCGGAGCTACTCAACAGCTACTTTGCCTCAGTCTTCACAAAAAAATTAAGCTGCAGCCAGACACCTAATAAGGATTATCTGGATAAGGAAGGGGGCAAGCTGAGGGANGAGATAACAAAAGAGACGGTCAGAGAGCTTTTGNTGGGTCTGAATGAATTCAAGTCAGCAGGGCCGGATGAACTTCATCCCAGGGTACTGAAGGAACTGGCAGAGGAGATCTCGGAGCCCTTGGCAATGATCTTCATGAAGTCGTGGGAGACGGGCCAGGTACCAGAGGACTGGAAAAGGGCCAACGTAGTGCCCCTCTTTAAAAAAGGCAAAAAGGAGGACCCGGGGAACTACAGACCGGTTAGCCTCACCTCAATACCTGGGAAGTTACTGGAGCATATCCTAAGGAAGGCCATTTGCAAGCATCTGGAGGAGGAAAGGCTGATCACAAGCAGCCAGCATGGATTTACGAAAAACAAATCGTGTCAAACAAACTTGATCTCCTTCTTTGACAAAGTAACTACTTGGGTGGATGAAGGGAATGCTGTGGATATAGTGTATCTGGACTTCAGCAAGGCTTTTGACAAGGTCCCGCATGACCTTCTCATAAACAAATTGGAGAAATGTGGGCTGGACAAAACTACTACAAGGTGGATAGATAACTGGCTCAATAGCCGCAAGCAAAGGGTAATTATTAACGGGTCCATGTCAGAATGGCAGGAGGTTTCGAGTGGAGTCCCACAGGGGTCTGTCCTGGGCCCGGTGCTGTTCAACGTGTTTATTAATGATTTGGATGCTGGCATAGAAAGCTTCTTGAGCAAGTTTGCAGACGACACGAAACTGGGAAGGATTGCGAACACGTTGGAGGACGNGAGCGCAATTCAGAAGGATCTCGACAGATTGGAAAGCTGGGCTAGAGCTAACAGGATGAAGTTCAATGCGGACAAATGCAAGGTGCTACACCTCGGGAGGAATAATCAAAAACACAAATACAAAATGGGTGACACCTGGCTGGATGGCAGCACTATGGAAAAGGACCTGGGAGTCTTGGTAGATCACAGACTCAATATGAGTCTGCAGTGTGATGCAGCCGCACAAAAGGCGAATGCGGTTTTGGGATGCATCAATAGAAGCATTAGGTGCAAGACACGGGAGGTGATAGTGCCTCTCTACTCGGCACTGGTTAGGCCTCATCTGGAGTACTGTGTGCAGTTCTGGGCTCCACATTTTAAAAAGGACGTGGAGAAGTTAGAGAGGGTCCAGAGGCGGGCAACAAAGATGATAAAGGGCTTGGAAGGCAAGTCATACGAGGAGAGGCTGAAGGAGCTAGGCATGTTCAGCTTGCGGAAAAGGCGCTTAAGAGGGGACATGATAGCAGTCTTCAAATACTTGAAGGGCTGCCATAAAGAAGAGGGAAAGCACCTTTTCTCTCTTGCTGCAGAGAGGAGGACGCGGACCAATGGCTTGAAGTTGCAGCAAAGTAGGTTTAGATTGGATATCCGGAAAAACTTCTTCACTGTTAGAACAGTGAGGCAGTGGAATAGACTGCCTAGGGAAGTTGTGGACTCTCCATCACTGGAGGTGTTCAAGAAGAGGTTGGACAGCCACTGGTCGGGGATGATCTAGGCGTAGCTATGCCTATGTTCTACTATGGGTATTTCCCATGCTTCTGGGCTTTGCTGGTTGCCCCCGCCTCCCCCCCCCCCCCTTTCTGCTACATGTGTCAGGGTGTTTTTAAGCCTTCCTCAGAGGCATTGGGTGTTGGCTGCAGCCAAGGCTGGGGACTTTGACTGGGGTGTGCCAATGCTCCTGCTGGGACCAAAGCCGGAGGTTCCTGGCTAGAGGGTCTTGCCCCTCCGCTCAGGGTCAGACCGATCGCCATATTTGGGGTCAGGAAGGAATTTTACCCCATGGTCAGATTGGTACGGACTGTGGGGGTTTTTGCCTTCCTCTGTAGCGGGGGGCGTGGCCCTCTACCCGGGATCTCTTGAGCATATATTGACAACATTTCATAGAAGCAGGACATTGGCTGCCGTGGTTCCCCTGCTTTACCTGTGGCAGGTTAGGGTGTTAGGTCTGTGTTGTGTCAGGGTTGACGGTAGTCTTACGTAGAGTTTAGATTATGGTTGTATGGGATGGTTTGGATAGGGATGATCCTGCCTCAGGCAGGGGGTTGGACTAGATGACCTCTGGAGGTCCCTTCCAGCCCTACTTCTCTATGATTCTATG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-3_Croc | KLF3 | 3338 | 3347 | - | 18.00 | GGCCACGCCC |
| CR1-3_Croc | Klf15 | 3062 | 3083 | + | 17.97 | GCCCCCGCCTCCCCCCCCCCCC |
| CR1-3_Croc | KLF11 | 3336 | 3345 | - | 17.92 | CCACGCCCCC |
| CR1-3_Croc | DOF3.2 | 1700 | 1715 | - | 17.70 | CTCCTTTTTGCCTTTT |
| CR1-3_Croc | SP8 | 3335 | 3345 | - | 17.62 | CCACGCCCCCC |
| CR1-3_Croc | Klf6-7-like | 3065 | 3084 | + | 17.60 | CCCGCCTCCCCCCCCCCCCC |
| CR1-3_Croc | ZNF740 | 3072 | 3081 | + | 17.57 | CCCCCCCCCC |
| CR1-3_Croc | ZNF740 | 3073 | 3082 | + | 17.57 | CCCCCCCCCC |
| CR1-3_Croc | ZNF740 | 3074 | 3083 | + | 17.57 | CCCCCCCCCC |
| CR1-3_Croc | ZNF740 | 3075 | 3084 | + | 17.57 | CCCCCCCCCC |
TFBS enrichment in GRCh38
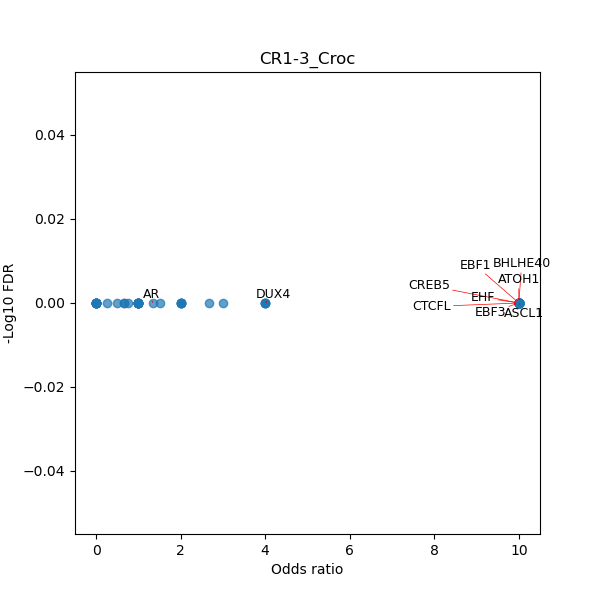
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.