CR1-3_Croc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001325 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 3605 |
| Kimura value | 33.02 |
| Tau index | 0.0000 |
| Description | CR1 (Chicken Repeat 1) retrotransposon, CR1-3_Croc subfamily |
| Comment | - |
| Sequence |
GGGATCCATCTGACGAGAACGGGGAAGAGCATCTTTGGGCGTCGTCTGGCCAACCTAGTAAGGAGGGCTTTAAACTAGGTTCCACGGGGATGGTGATCAAAACCCTCCAGCAACAGAGCACACAGTCCAGAGGGGAAAGGACTCGAAGAAGCAACTCAACTCTGGGAAAGGGCCAGAGAGCTACAGGACTGCAACAAGTGGCCAAAAGGAAGCGAGTAGGTCAAAATGCAAAATACGTCCGATGCATGTATACAAATGCGAGAAGTATGGGGAATAAACAAGATGAGCTGGAAGCTGCAGCCTGTGAAAAGAACTATGATCTGATTGGCATCACAGAGACCTGGTGGGACAGCTCTTATGACTGGAATGTCAACATCGAGGGTTACACTCTGTTTCGGAAGGACAGGGTTGGGAGAAAAGGTGGTGGTGTTGCCTTGTATGTCAAAAACACATACACTTGTTCCCTGGTTCAGGAGGAAGAAGACAGCAAAGCAGAATGCATTTGGGTGAAGATGAAAGGTGAAAGATGCAGCACCGATATAATGGTGGGGATTTATTATAGGCCACCAAATCAGGAAGAGGAGGTGGATGAGGCACTCTTCAAGCAGGTGACAAATCTATCCAAAAGCTATGGGATGGTACTGATGGGAGACTTCAATTTTCCGGACATCTGCTGGAAGAACAATGCAGCCAAACATAAAATGTCAAGCCGGTTCCTAACTTGTGTGGAGGACAACTTTCTGTTCCAGAAAGTGGAGGATACAACTAGGGGATCATCTATCTTGGATCTTGTCCTGACCAACAGGGAAGAACTGGTGGAGGATGTGAAGGTGATGGGTAACTTGGGAGGAAGCGATCACGATATGATAGAATTCCAGATCCTGAGACGGGGAAAGCATGAGGTCAGCAAAATTAGGACGCTAGATTTCAAAAAGGCGGATTTCATCCGACTCAAGGAGTTAATAGGCATGGTGCCATGGGAGGACAGACTGAAGGATAAAGGCGCCCAGGAGGGCTGGGAGCTTCTAAAGGGCACAGTATTGGAAGCCCAACAAGAAACTGTCCCGACACGATGGAAGCACAAGAAGAATGGCAAGAGACCAATGTGGCTGCACAAGGAGCTTCTAAAATGCCTCAAATGCAAAAGGAAAGCATACAGGCAATGGAAGAATGGGCAGGTCACCAAGGAAGCGTACCAGGAAATAGCAAGAACCTGCAGGGACAAAATCAGGAAGGCAAAGATAAAGAACGAGCTGCACCTGGCGAAGGAGGTTAAGGACAACAAGAAGAGGTTCTATAAGTATGTTGGCCAGAAAAGAAGGACCAAGGAAGCTGTGGGTCCNCTGTTAACAAGCCAGGGGGAACTCCTAACNGAAGATGCAAAGAAGGCGGAGCTACTCAACAGCTACTTTGCCTCAGTCTTCACAAAAAAATTAAGCTGCAGCCAGACACCTAATAAGGATTATCTGGATAAGGAAGGGGGCAAGCTGAGGGANGAGATAACAAAAGAGACGGTCAGAGAGCTTTTGNTGGGTCTGAATGAATTCAAGTCAGCAGGGCCGGATGAACTTCATCCCAGGGTACTGAAGGAACTGGCAGAGGAGATCTCGGAGCCCTTGGCAATGATCTTCATGAAGTCGTGGGAGACGGGCCAGGTACCAGAGGACTGGAAAAGGGCCAACGTAGTGCCCCTCTTTAAAAAAGGCAAAAAGGAGGACCCGGGGAACTACAGACCGGTTAGCCTCACCTCAATACCTGGGAAGTTACTGGAGCATATCCTAAGGAAGGCCATTTGCAAGCATCTGGAGGAGGAAAGGCTGATCACAAGCAGCCAGCATGGATTTACGAAAAACAAATCGTGTCAAACAAACTTGATCTCCTTCTTTGACAAAGTAACTACTTGGGTGGATGAAGGGAATGCTGTGGATATAGTGTATCTGGACTTCAGCAAGGCTTTTGACAAGGTCCCGCATGACCTTCTCATAAACAAATTGGAGAAATGTGGGCTGGACAAAACTACTACAAGGTGGATAGATAACTGGCTCAATAGCCGCAAGCAAAGGGTAATTATTAACGGGTCCATGTCAGAATGGCAGGAGGTTTCGAGTGGAGTCCCACAGGGGTCTGTCCTGGGCCCGGTGCTGTTCAACGTGTTTATTAATGATTTGGATGCTGGCATAGAAAGCTTCTTGAGCAAGTTTGCAGACGACACGAAACTGGGAAGGATTGCGAACACGTTGGAGGACGNGAGCGCAATTCAGAAGGATCTCGACAGATTGGAAAGCTGGGCTAGAGCTAACAGGATGAAGTTCAATGCGGACAAATGCAAGGTGCTACACCTCGGGAGGAATAATCAAAAACACAAATACAAAATGGGTGACACCTGGCTGGATGGCAGCACTATGGAAAAGGACCTGGGAGTCTTGGTAGATCACAGACTCAATATGAGTCTGCAGTGTGATGCAGCCGCACAAAAGGCGAATGCGGTTTTGGGATGCATCAATAGAAGCATTAGGTGCAAGACACGGGAGGTGATAGTGCCTCTCTACTCGGCACTGGTTAGGCCTCATCTGGAGTACTGTGTGCAGTTCTGGGCTCCACATTTTAAAAAGGACGTGGAGAAGTTAGAGAGGGTCCAGAGGCGGGCAACAAAGATGATAAAGGGCTTGGAAGGCAAGTCATACGAGGAGAGGCTGAAGGAGCTAGGCATGTTCAGCTTGCGGAAAAGGCGCTTAAGAGGGGACATGATAGCAGTCTTCAAATACTTGAAGGGCTGCCATAAAGAAGAGGGAAAGCACCTTTTCTCTCTTGCTGCAGAGAGGAGGACGCGGACCAATGGCTTGAAGTTGCAGCAAAGTAGGTTTAGATTGGATATCCGGAAAAACTTCTTCACTGTTAGAACAGTGAGGCAGTGGAATAGACTGCCTAGGGAAGTTGTGGACTCTCCATCACTGGAGGTGTTCAAGAAGAGGTTGGACAGCCACTGGTCGGGGATGATCTAGGCGTAGCTATGCCTATGTTCTACTATGGGTATTTCCCATGCTTCTGGGCTTTGCTGGTTGCCCCCGCCTCCCCCCCCCCCCCTTTCTGCTACATGTGTCAGGGTGTTTTTAAGCCTTCCTCAGAGGCATTGGGTGTTGGCTGCAGCCAAGGCTGGGGACTTTGACTGGGGTGTGCCAATGCTCCTGCTGGGACCAAAGCCGGAGGTTCCTGGCTAGAGGGTCTTGCCCCTCCGCTCAGGGTCAGACCGATCGCCATATTTGGGGTCAGGAAGGAATTTTACCCCATGGTCAGATTGGTACGGACTGTGGGGGTTTTTGCCTTCCTCTGTAGCGGGGGGCGTGGCCCTCTACCCGGGATCTCTTGAGCATATATTGACAACATTTCATAGAAGCAGGACATTGGCTGCCGTGGTTCCCCTGCTTTACCTGTGGCAGGTTAGGGTGTTAGGTCTGTGTTGTGTCAGGGTTGACGGTAGTCTTACGTAGAGTTTAGATTATGGTTGTATGGGATGGTTTGGATAGGGATGATCCTGCCTCAGGCAGGGGGTTGGACTAGATGACCTCTGGAGGTCCCTTCCAGCCCTACTTCTCTATGATTCTATG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-3_Croc | Klf15 | 3063 | 3084 | + | 25.20 | CCCCCGCCTCCCCCCCCCCCCC |
| CR1-3_Croc | PRDM9 | 3066 | 3085 | - | 21.01 | AGGGGGGGGGGGGGAGGCGG |
| CR1-3_Croc | CG3065 | 3336 | 3346 | - | 20.72 | GCCACGCCCCC |
| CR1-3_Croc | PRDM9 | 3063 | 3082 | - | 19.87 | GGGGGGGGGGGAGGCGGGGG |
| CR1-3_Croc | SP3 | 3336 | 3346 | - | 19.87 | GCCACGCCCCC |
| CR1-3_Croc | KLF16 | 3336 | 3346 | - | 19.28 | GCCACGCCCCC |
| CR1-3_Croc | Klf15 | 3064 | 3085 | + | 18.95 | CCCCGCCTCCCCCCCCCCCCCT |
| CR1-3_Croc | Spps | 3336 | 3346 | - | 18.44 | GCCACGCCCCC |
| CR1-3_Croc | MTF1 | 2199 | 2212 | + | 18.24 | TTTGCAGACGACAC |
| CR1-3_Croc | PRDM9 | 3060 | 3079 | - | 18.07 | GGGGGGGGAGGCGGGGGCAA |
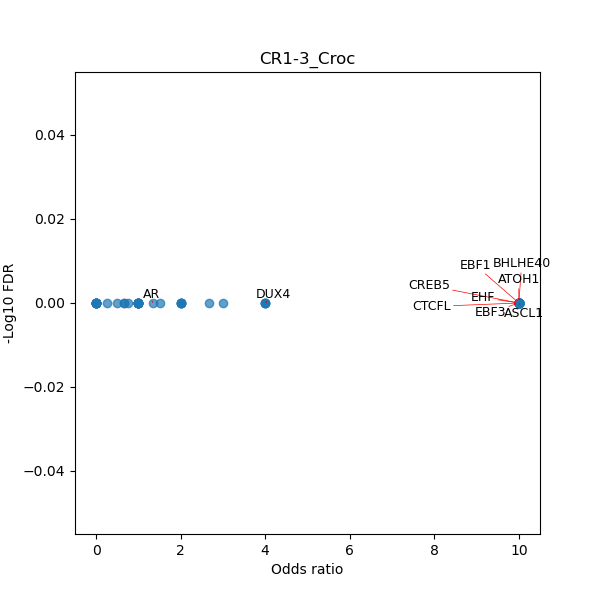
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.