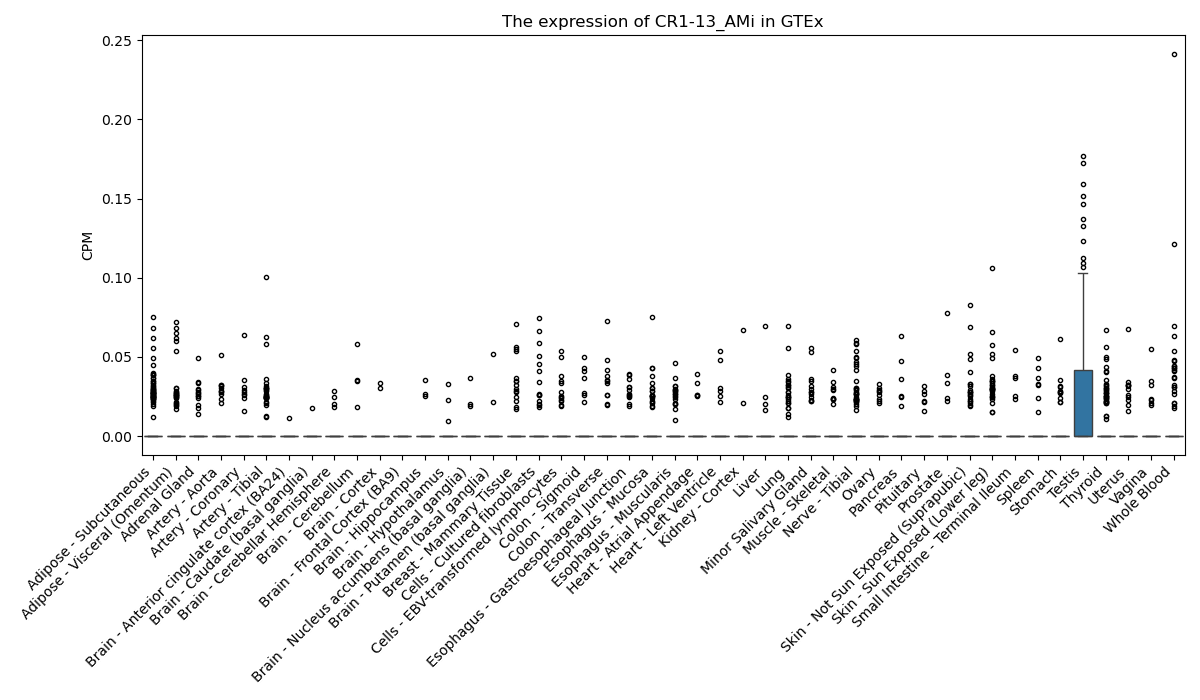
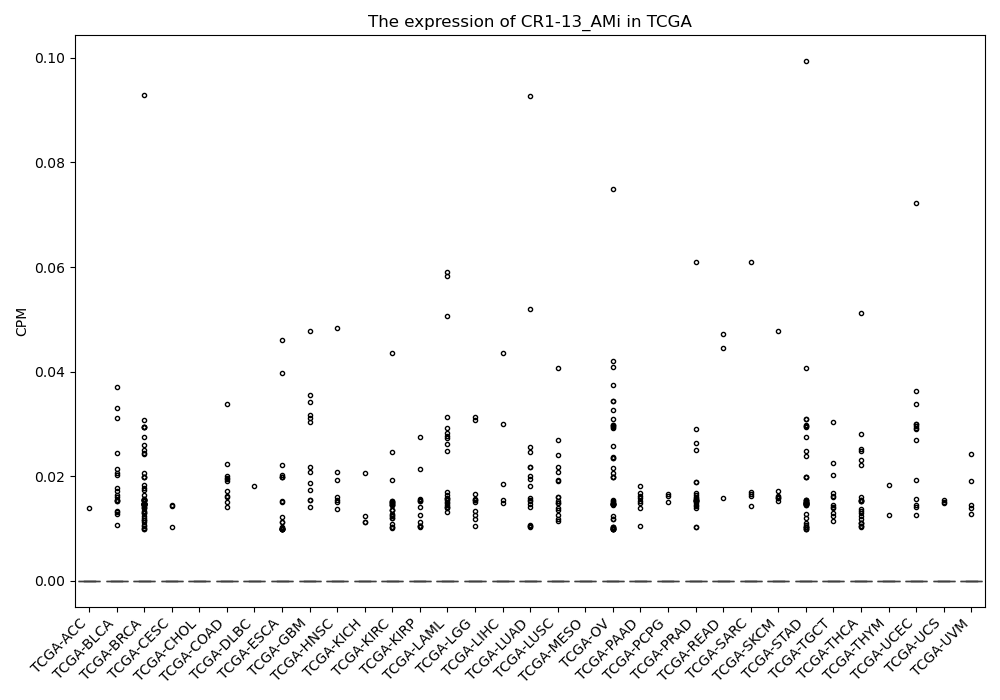
CR1-13_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001291 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 724 |
| Kimura value | 35.73 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAGGGATTAGAAAATAATACTGAANATANTATAATGCCATTATATAAATCGATGGTGCACTTCACTTGAATACCGCGTTCAGTTCTGGTCACCCTATCTCAAAGAAATANAGCAGAAATAAGGAGTTCGAGACGGGCGACGAAAATGATTAGAGGCAGAGAGACTTCCATATGAAGAGAGANTGAAGACTGGACTGTTTAGTTTAGAGAGGAGACGAATGAGGGGACATGATAGAGGTATACAAAATAATGAATGGACAGGGGAGCAATCGGGAGCTCCTATTTACCCTTTCTCATAATACAAGAACAAGGACATTCAATGAAATTGAAAGGCAACAATTTAAAACTGATAAAAGGAAATACTTTTTTTACACAATGCATAATTAACCTGTGGAACTCACTGCCACAAGATATCACTGAGGCCAAGAGCTTAGCAGGATTCAAAAAGGATTAGACATTTATATGGATAACGAGAACATCCGCAGTTACATTAGATAGGATAAAAAATATAAGGGATATAANCCTCATGCTTCAGGGCATAAGCCAACCACTAACTGACGGGGTTAGGAAGAAACTTCCCCTATGGGCAGGTTATTCCATAATTGCCATTATGGGGNTTTTGCACCTTCCTCTGAAGCATCTGGTACTGGCCACTGTCAGAGACAGGATACTGGACTAGATGGACCACTGGTCATTCCACTATNGCGAATTAATTGTTCTTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-13_AMi | Stat6 | 629 | 638 | + | 13.48 | TTCCTCTGAA |
| CR1-13_AMi | PK10344.1 | 683 | 690 | + | 13.46 | TGGACCAC |
| CR1-13_AMi | TFAP2A | 579 | 591 | - | 13.41 | TGCCCATAGGGGA |
| CR1-13_AMi | PITX2 | 6 | 13 | - | 13.40 | CTAATCCC |
| CR1-13_AMi | ROX1 | 712 | 723 | + | 13.39 | TTAATTGTTCTT |
| CR1-13_AMi | HAT2 | 147 | 154 | - | 13.38 | TAATCATT |
| CR1-13_AMi | ceh-18 | 379 | 386 | + | 13.35 | TGCATAAT |
| CR1-13_AMi | TFAP2A | 579 | 591 | + | 13.34 | TCCCCTATGGGCA |
| CR1-13_AMi | HSFB4 | 571 | 580 | - | 13.30 | GAAGTTTCTT |
| CR1-13_AMi | GRF6 | 657 | 663 | - | 13.30 | TCTGACA |
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.