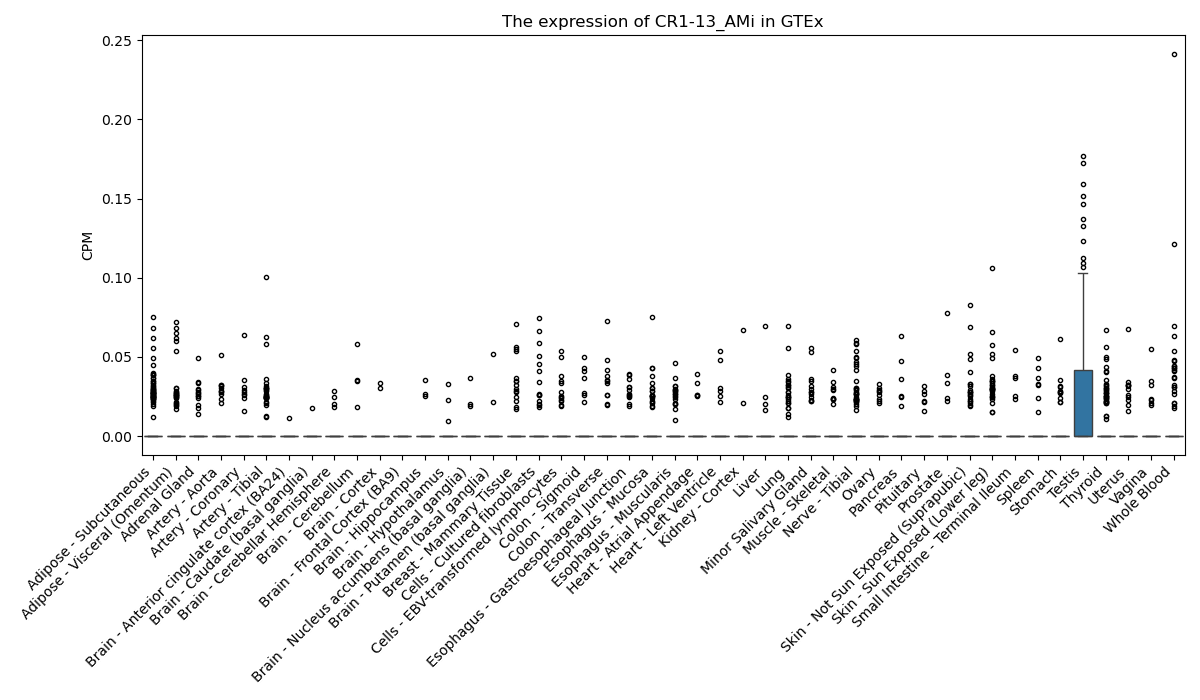
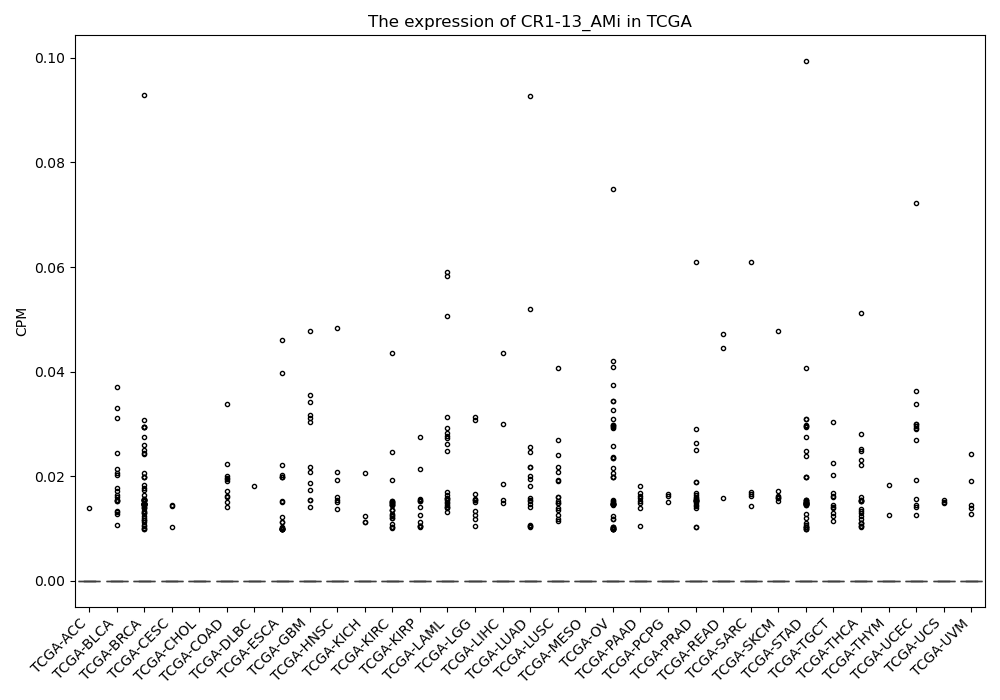
CR1-13_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001291 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 724 |
| Kimura value | 35.73 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAGGGATTAGAAAATAATACTGAANATANTATAATGCCATTATATAAATCGATGGTGCACTTCACTTGAATACCGCGTTCAGTTCTGGTCACCCTATCTCAAAGAAATANAGCAGAAATAAGGAGTTCGAGACGGGCGACGAAAATGATTAGAGGCAGAGAGACTTCCATATGAAGAGAGANTGAAGACTGGACTGTTTAGTTTAGAGAGGAGACGAATGAGGGGACATGATAGAGGTATACAAAATAATGAATGGACAGGGGAGCAATCGGGAGCTCCTATTTACCCTTTCTCATAATACAAGAACAAGGACATTCAATGAAATTGAAAGGCAACAATTTAAAACTGATAAAAGGAAATACTTTTTTTACACAATGCATAATTAACCTGTGGAACTCACTGCCACAAGATATCACTGAGGCCAAGAGCTTAGCAGGATTCAAAAAGGATTAGACATTTATATGGATAACGAGAACATCCGCAGTTACATTAGATAGGATAAAAAATATAAGGGATATAANCCTCATGCTTCAGGGCATAAGCCAACCACTAACTGACGGGGTTAGGAAGAAACTTCCCCTATGGGCAGGTTATTCCATAATTGCCATTATGGGGNTTTTGCACCTTCCTCTGAAGCATCTGGTACTGGCCACTGTCAGAGACAGGATACTGGACTAGATGGACCACTGGTCATTCCACTATNGCGAATTAATTGTTCTTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-13_AMi | LFY | 681 | 699 | - | 17.65 | GAATGACCAGTGGTCCATC |
| CR1-13_AMi | LFY | 681 | 699 | + | 17.65 | GATGGACCACTGGTCATTC |
| CR1-13_AMi | ZNF530 | 254 | 267 | + | 17.09 | GAATGGACAGGGGA |
| CR1-13_AMi | MYB30 | 546 | 557 | + | 16.15 | CCAACCACTAAC |
| CR1-13_AMi | eor-1 | 154 | 166 | + | 15.50 | AGAGGCAGAGAGA |
| CR1-13_AMi | ZNF816 | 224 | 238 | + | 15.37 | AGGGGACATGATAGA |
| CR1-13_AMi | POU4F1 | 379 | 390 | + | 15.16 | TGCATAATTAAC |
| CR1-13_AMi | POU4F3 | 379 | 390 | + | 15.14 | TGCATAATTAAC |
| CR1-13_AMi | AT1G19000 | 463 | 475 | + | 15.09 | ATATGGATAACGA |
| CR1-13_AMi | SoxN | 320 | 330 | - | 15.07 | CAATTTCATTG |
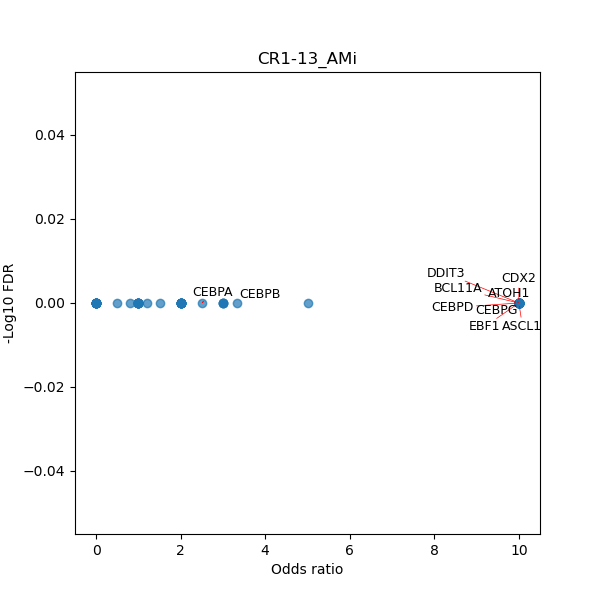
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.