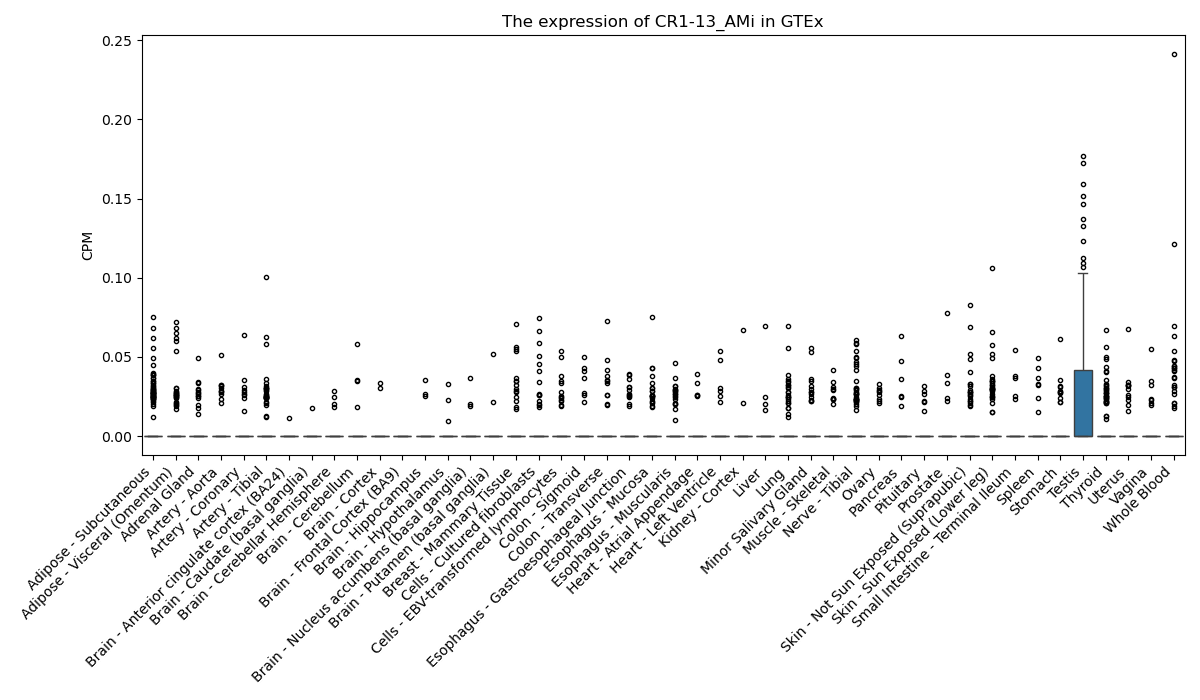
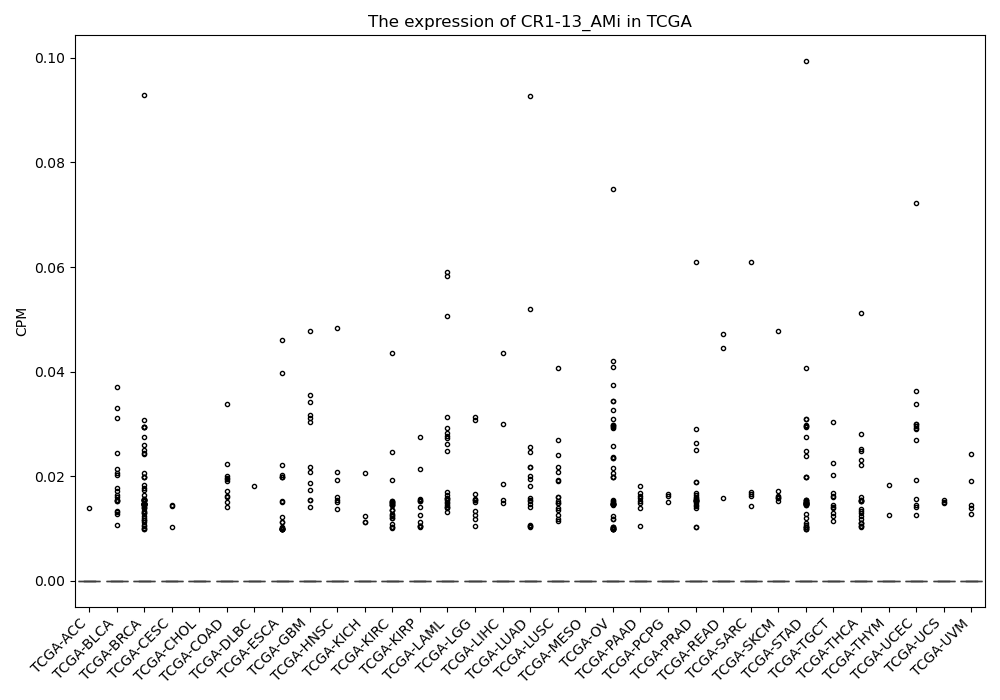
CR1-13_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001291 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 724 |
| Kimura value | 35.73 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAGGGATTAGAAAATAATACTGAANATANTATAATGCCATTATATAAATCGATGGTGCACTTCACTTGAATACCGCGTTCAGTTCTGGTCACCCTATCTCAAAGAAATANAGCAGAAATAAGGAGTTCGAGACGGGCGACGAAAATGATTAGAGGCAGAGAGACTTCCATATGAAGAGAGANTGAAGACTGGACTGTTTAGTTTAGAGAGGAGACGAATGAGGGGACATGATAGAGGTATACAAAATAATGAATGGACAGGGGAGCAATCGGGAGCTCCTATTTACCCTTTCTCATAATACAAGAACAAGGACATTCAATGAAATTGAAAGGCAACAATTTAAAACTGATAAAAGGAAATACTTTTTTTACACAATGCATAATTAACCTGTGGAACTCACTGCCACAAGATATCACTGAGGCCAAGAGCTTAGCAGGATTCAAAAAGGATTAGACATTTATATGGATAACGAGAACATCCGCAGTTACATTAGATAGGATAAAAAATATAAGGGATATAANCCTCATGCTTCAGGGCATAAGCCAACCACTAACTGACGGGGTTAGGAAGAAACTTCCCCTATGGGCAGGTTATTCCATAATTGCCATTATGGGGNTTTTGCACCTTCCTCTGAAGCATCTGGTACTGGCCACTGTCAGAGACAGGATACTGGACTAGATGGACCACTGGTCATTCCACTATNGCGAATTAATTGTTCTTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-13_AMi | MYB60 | 546 | 555 | + | 14.16 | CCAACCACTA |
| CR1-13_AMi | HSFB2A | 572 | 581 | - | 13.96 | GGAAGTTTCT |
| CR1-13_AMi | Irf1 | 323 | 333 | + | 13.90 | TGAAATTGAAA |
| CR1-13_AMi | pqm-1 | 348 | 355 | + | 13.89 | ACTGATAA |
| CR1-13_AMi | DOF5.1 | 1 | 19 | + | 13.85 | AAAAAGGGATTAGAAAATA |
| CR1-13_AMi | ces-2 | 42 | 50 | + | 13.58 | ATTATATAA |
| CR1-13_AMi | DOF3.4 | 1 | 17 | - | 13.53 | TTTTCTAATCCCTTTTT |
| CR1-13_AMi | egl-27 | 347 | 357 | - | 13.53 | TTTTATCAGTT |
| CR1-13_AMi | blmp-1 | 324 | 334 | + | 13.51 | GAAATTGAAAG |
| CR1-13_AMi | HSFB2B | 571 | 581 | - | 13.50 | GGAAGTTTCTT |
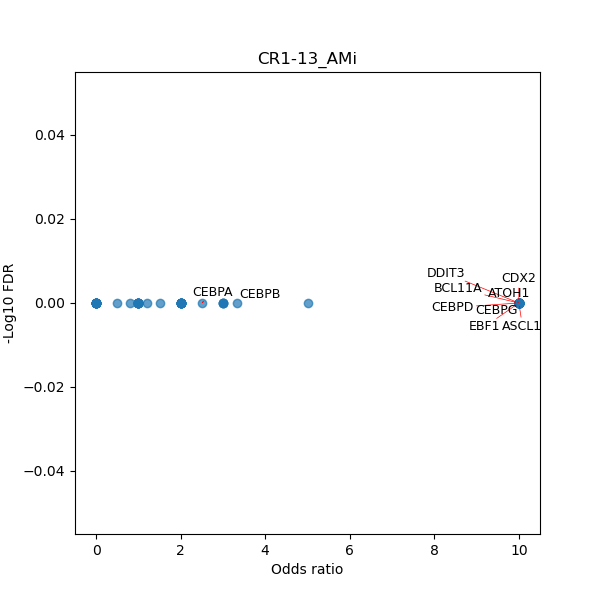
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.