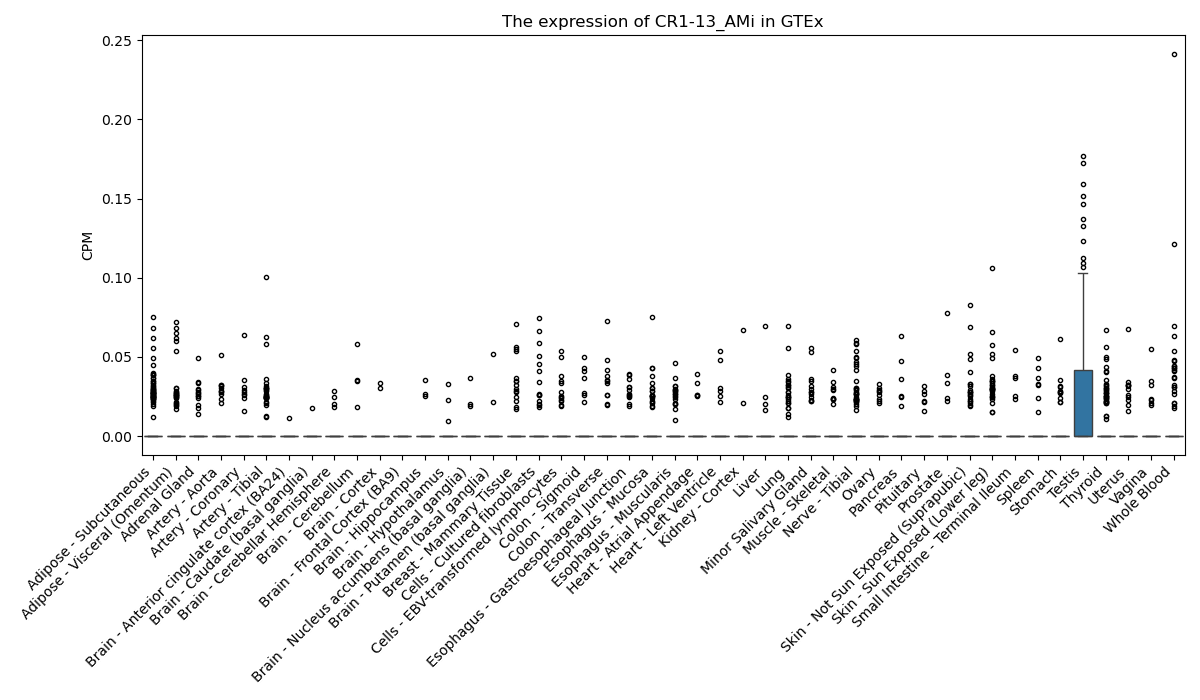
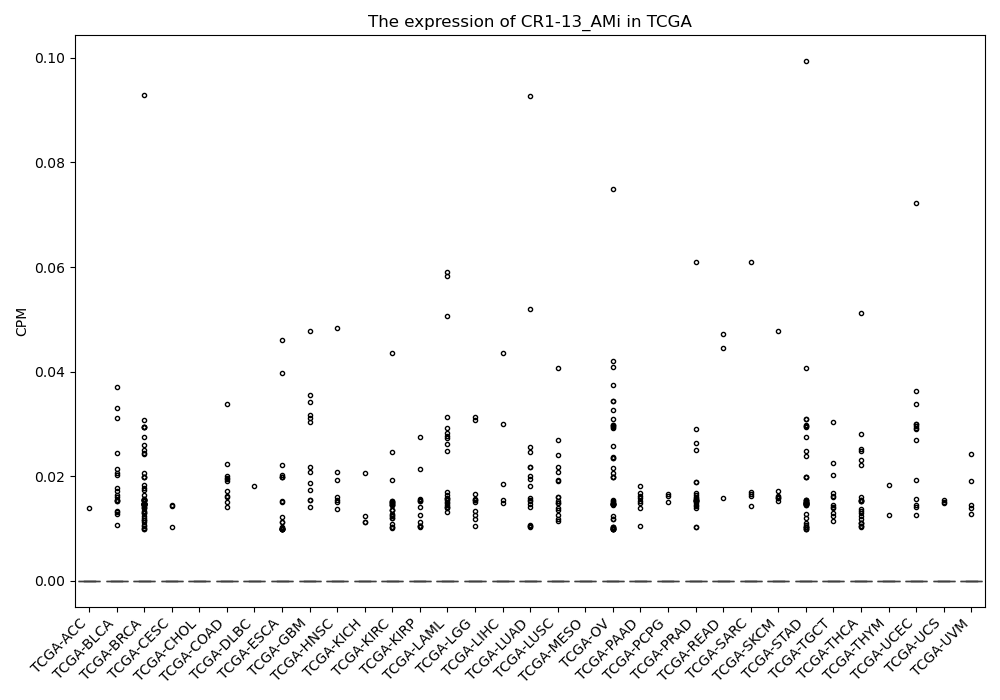
CR1-13_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001291 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 724 |
| Kimura value | 35.73 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAGGGATTAGAAAATAATACTGAANATANTATAATGCCATTATATAAATCGATGGTGCACTTCACTTGAATACCGCGTTCAGTTCTGGTCACCCTATCTCAAAGAAATANAGCAGAAATAAGGAGTTCGAGACGGGCGACGAAAATGATTAGAGGCAGAGAGACTTCCATATGAAGAGAGANTGAAGACTGGACTGTTTAGTTTAGAGAGGAGACGAATGAGGGGACATGATAGAGGTATACAAAATAATGAATGGACAGGGGAGCAATCGGGAGCTCCTATTTACCCTTTCTCATAATACAAGAACAAGGACATTCAATGAAATTGAAAGGCAACAATTTAAAACTGATAAAAGGAAATACTTTTTTTACACAATGCATAATTAACCTGTGGAACTCACTGCCACAAGATATCACTGAGGCCAAGAGCTTAGCAGGATTCAAAAAGGATTAGACATTTATATGGATAACGAGAACATCCGCAGTTACATTAGATAGGATAAAAAATATAAGGGATATAANCCTCATGCTTCAGGGCATAAGCCAACCACTAACTGACGGGGTTAGGAAGAAACTTCCCCTATGGGCAGGTTATTCCATAATTGCCATTATGGGGNTTTTGCACCTTCCTCTGAAGCATCTGGTACTGGCCACTGTCAGAGACAGGATACTGGACTAGATGGACCACTGGTCATTCCACTATNGCGAATTAATTGTTCTTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-13_AMi | Mecom | 495 | 505 | + | 14.75 | AGATAGGATAA |
| CR1-13_AMi | Zm00001d038683 | 683 | 690 | - | 14.75 | GTGGTCCA |
| CR1-13_AMi | NTL8 | 570 | 577 | + | 14.41 | GAAGAAAC |
| CR1-13_AMi | MYB94 | 546 | 555 | + | 14.38 | CCAACCACTA |
| CR1-13_AMi | AT1G74840 | 464 | 472 | + | 14.36 | TATGGATAA |
| CR1-13_AMi | EIL4 | 318 | 326 | + | 14.36 | TTCAATGAA |
| CR1-13_AMi | ATMYB31 | 547 | 561 | + | 14.31 | CAACCACTAACTGAC |
| CR1-13_AMi | MYB31 | 547 | 561 | + | 14.31 | CAACCACTAACTGAC |
| CR1-13_AMi | DOF5.8 | 1 | 19 | - | 14.27 | TATTTTCTAATCCCTTTTT |
| CR1-13_AMi | HSFA1B | 573 | 580 | + | 14.20 | GAAACTTC |
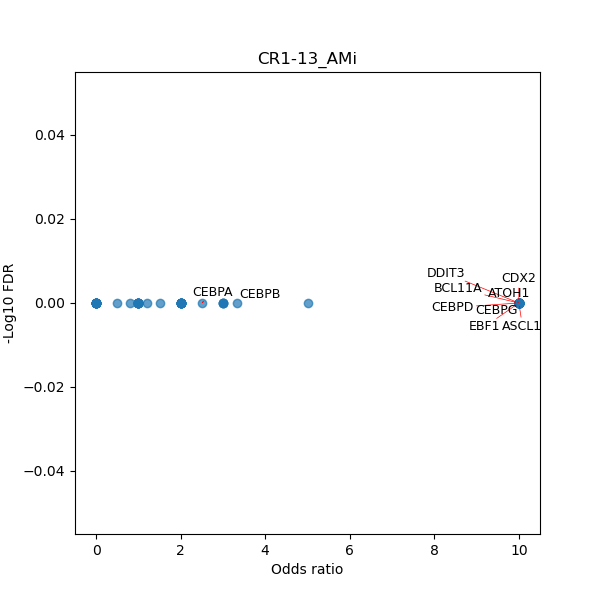
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.