CR1-12_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001300 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 614 |
| Kimura value | 33.40 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAAGGAANCTTACNAGAAATGGAAANTTGGGNAAACAANCAGGGAGGAGTATAAGGGCATTGCTAAGGCTTGCAGGGAGAAAGTAGGAAGGCAAAACGCAATGAGTTAATGCTAGCCAAAGGGGTTAAGGGGAATAAGAAGGGCTTTTACAAATATATCAGGGGGAAAAGAAATACTAGAGAGAAAGTAGGTCCATTAATAAATGAGGGAGGGAATTAGTAATGATGACTCTAAAATGGCTGAGATACTGAACTGCTTTTTAAATCTGTCTTTAACAAGAAAAAGAAAAGAATGGGCAGACAATTAGGAAGGAGGTGTAAAACAGCTATTGATAAAGAGCAGGTAAGGGAATACTTGGAAAACTGAACGTATACAAATCAGCGGGTCCGGATGACATGCATCCCAGGGTGCTGAGGGAATTAGCTAATGAACTACNGAACCACTGGCAATTATTTTTGAAAAGTCATGGAGAACTGGGGAGGTCCAGAGGACTGGAGAAAAGCAAATGTAGTACCNATCTTCAAAAAAGGGAAGAAGAGNGATCCNGGCAATTACAGCCAGTCAGTCTNACCTCAATCCCTGGNAAAATAATGGAACAAATATTAAAGGAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-12_AMi | ATHB-51 | 450 | 457 | - | 12.90 | AATAATTG |
| CR1-12_AMi | PI | 525 | 538 | + | 12.83 | CAAAAAAGGGAAGA |
| CR1-12_AMi | ANL2 | 200 | 209 | - | 12.83 | TCATTTATTA |
| CR1-12_AMi | CAD1 | 218 | 228 | - | 12.82 | TCATTACTAAT |
| CR1-12_AMi | HAT22 | 450 | 458 | - | 12.81 | AAATAATTG |
| CR1-12_AMi | DOF3.4 | 283 | 299 | - | 12.77 | CCCATTCTTTTCTTTTT |
| CR1-12_AMi | YAP7 | 218 | 228 | + | 12.74 | ATTAGTAATGA |
| CR1-12_AMi | ZNF135 | 478 | 491 | - | 12.70 | TCTGGACCTCCCCA |
| CR1-12_AMi | ATHB-X | 450 | 458 | + | 12.70 | CAATTATTT |
| CR1-12_AMi | FoxA-b | 600 | 607 | - | 12.69 | AATATTTG |
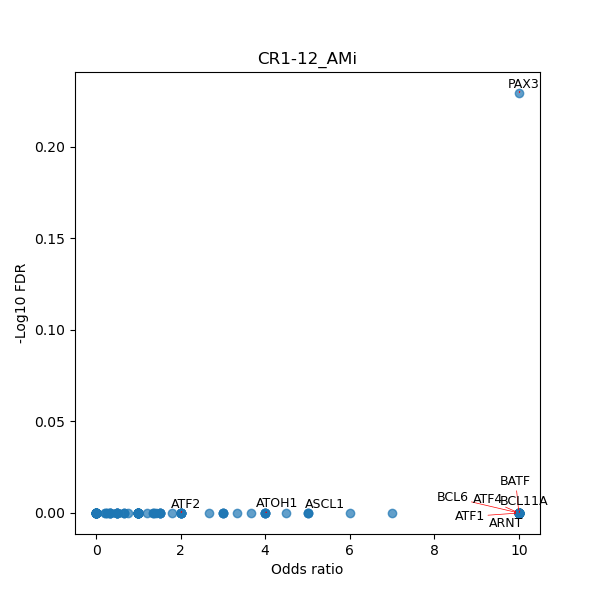
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.