CR1-12_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001300 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 614 |
| Kimura value | 33.40 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAAGGAANCTTACNAGAAATGGAAANTTGGGNAAACAANCAGGGAGGAGTATAAGGGCATTGCTAAGGCTTGCAGGGAGAAAGTAGGAAGGCAAAACGCAATGAGTTAATGCTAGCCAAAGGGGTTAAGGGGAATAAGAAGGGCTTTTACAAATATATCAGGGGGAAAAGAAATACTAGAGAGAAAGTAGGTCCATTAATAAATGAGGGAGGGAATTAGTAATGATGACTCTAAAATGGCTGAGATACTGAACTGCTTTTTAAATCTGTCTTTAACAAGAAAAAGAAAAGAATGGGCAGACAATTAGGAAGGAGGTGTAAAACAGCTATTGATAAAGAGCAGGTAAGGGAATACTTGGAAAACTGAACGTATACAAATCAGCGGGTCCGGATGACATGCATCCCAGGGTGCTGAGGGAATTAGCTAATGAACTACNGAACCACTGGCAATTATTTTTGAAAAGTCATGGAGAACTGGGGAGGTCCAGAGGACTGGAGAAAAGCAAATGTAGTACCNATCTTCAAAAAAGGGAAGAAGAGNGATCCNGGCAATTACAGCCAGTCAGTCTNACCTCAATCCCTGGNAAAATAATGGAACAAATATTAAAGGAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-12_AMi | REV | 220 | 233 | - | 16.58 | AGTCATCATTACTA |
| CR1-12_AMi | ATHB-15 | 219 | 231 | + | 16.48 | TTAGTAATGATGA |
| CR1-12_AMi | DOF3.6 | 277 | 297 | - | 15.92 | CATTCTTTTCTTTTTCTTGTT |
| CR1-12_AMi | ken | 79 | 87 | - | 14.72 | ACTTTCTCC |
| CR1-12_AMi | CDF5 | 277 | 297 | - | 14.55 | CATTCTTTTCTTTTTCTTGTT |
| CR1-12_AMi | dsf | 462 | 469 | + | 14.52 | AAAAGTCA |
| CR1-12_AMi | PI | 279 | 292 | + | 14.42 | CAAGAAAAAGAAAA |
| CR1-12_AMi | AZF1 | 283 | 291 | + | 14.41 | AAAAAGAAA |
| CR1-12_AMi | blmp-1 | 282 | 292 | + | 14.20 | GAAAAAGAAAA |
| CR1-12_AMi | ken | 183 | 191 | - | 14.08 | ACTTTCTCT |
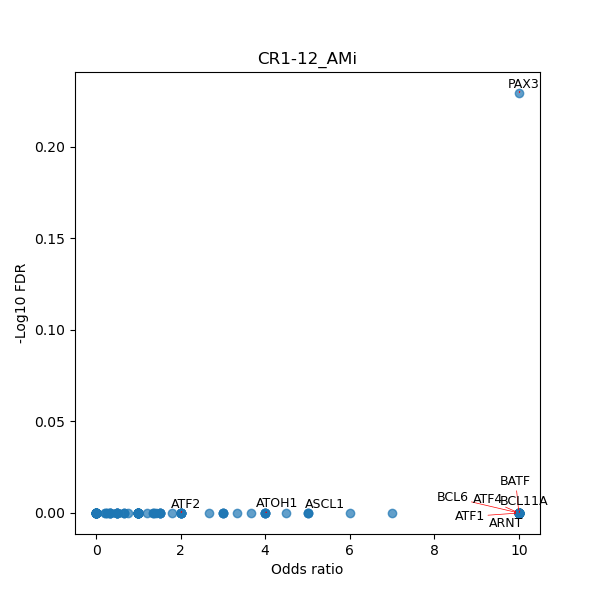
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.