CR1-12_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001300 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 614 |
| Kimura value | 33.40 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAAGGAANCTTACNAGAAATGGAAANTTGGGNAAACAANCAGGGAGGAGTATAAGGGCATTGCTAAGGCTTGCAGGGAGAAAGTAGGAAGGCAAAACGCAATGAGTTAATGCTAGCCAAAGGGGTTAAGGGGAATAAGAAGGGCTTTTACAAATATATCAGGGGGAAAAGAAATACTAGAGAGAAAGTAGGTCCATTAATAAATGAGGGAGGGAATTAGTAATGATGACTCTAAAATGGCTGAGATACTGAACTGCTTTTTAAATCTGTCTTTAACAAGAAAAAGAAAAGAATGGGCAGACAATTAGGAAGGAGGTGTAAAACAGCTATTGATAAAGAGCAGGTAAGGGAATACTTGGAAAACTGAACGTATACAAATCAGCGGGTCCGGATGACATGCATCCCAGGGTGCTGAGGGAATTAGCTAATGAACTACNGAACCACTGGCAATTATTTTTGAAAAGTCATGGAGAACTGGGGAGGTCCAGAGGACTGGAGAAAAGCAAATGTAGTACCNATCTTCAAAAAAGGGAAGAAGAGNGATCCNGGCAATTACAGCCAGTCAGTCTNACCTCAATCCCTGGNAAAATAATGGAACAAATATTAAAGGAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-12_AMi | Prdm5 | 467 | 477 | - | 13.37 | GTTCTCCATGA |
| CR1-12_AMi | DOF5.1 | 283 | 301 | + | 13.30 | AAAAAGAAAAGAATGGGCA |
| CR1-12_AMi | PRDM1 | 184 | 190 | - | 13.28 | CTTTCTC |
| CR1-12_AMi | PRDM1 | 80 | 86 | - | 13.28 | CTTTCTC |
| CR1-12_AMi | RELB | 132 | 138 | - | 13.14 | ATTCCCC |
| CR1-12_AMi | ZHD3 | 450 | 458 | + | 13.12 | CAATTATTT |
| CR1-12_AMi | ZFP42 | 235 | 247 | + | 13.11 | TAAAATGGCTGAG |
| CR1-12_AMi | NAC053 | 279 | 285 | + | 13.01 | CAAGAAA |
| CR1-12_AMi | HDG1 | 200 | 209 | + | 13.00 | TAATAAATGA |
| CR1-12_AMi | NAC078 | 279 | 285 | + | 12.96 | CAAGAAA |
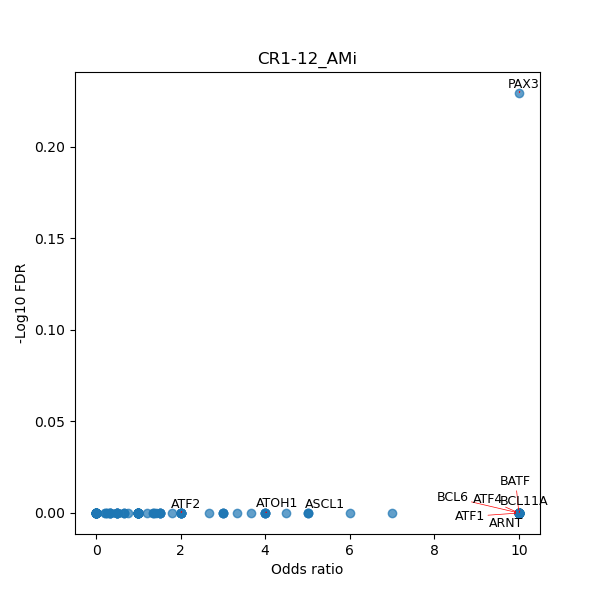
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.