CR1-12_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001300 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 614 |
| Kimura value | 33.40 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | Fragment. |
| Sequence |
AAAAAAGGAANCTTACNAGAAATGGAAANTTGGGNAAACAANCAGGGAGGAGTATAAGGGCATTGCTAAGGCTTGCAGGGAGAAAGTAGGAAGGCAAAACGCAATGAGTTAATGCTAGCCAAAGGGGTTAAGGGGAATAAGAAGGGCTTTTACAAATATATCAGGGGGAAAAGAAATACTAGAGAGAAAGTAGGTCCATTAATAAATGAGGGAGGGAATTAGTAATGATGACTCTAAAATGGCTGAGATACTGAACTGCTTTTTAAATCTGTCTTTAACAAGAAAAAGAAAAGAATGGGCAGACAATTAGGAAGGAGGTGTAAAACAGCTATTGATAAAGAGCAGGTAAGGGAATACTTGGAAAACTGAACGTATACAAATCAGCGGGTCCGGATGACATGCATCCCAGGGTGCTGAGGGAATTAGCTAATGAACTACNGAACCACTGGCAATTATTTTTGAAAAGTCATGGAGAACTGGGGAGGTCCAGAGGACTGGAGAAAAGCAAATGTAGTACCNATCTTCAAAAAAGGGAAGAAGAGNGATCCNGGCAATTACAGCCAGTCAGTCTNACCTCAATCCCTGGNAAAATAATGGAACAAATATTAAAGGAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-12_AMi | Nfatc2 | 21 | 28 | + | 14.03 | AATGGAAA |
| CR1-12_AMi | SPIC | 527 | 539 | + | 13.97 | AAAAAGGGAAGAA |
| CR1-12_AMi | ZNF263 | 45 | 51 | + | 13.83 | GGGAGGA |
| CR1-12_AMi | FUS3 | 397 | 403 | + | 13.75 | ACATGCA |
| CR1-12_AMi | Zm00001d027846 | 285 | 292 | + | 13.75 | AAAGAAAA |
| CR1-12_AMi | USV1 | 160 | 169 | - | 13.67 | TCCCCCTGAT |
| CR1-12_AMi | ATHB-53 | 450 | 458 | - | 13.67 | AAATAATTG |
| CR1-12_AMi | Kr | 122 | 130 | - | 13.61 | TAACCCCTT |
| CR1-12_AMi | Stat2 | 282 | 291 | + | 13.53 | GAAAAAGAAA |
| CR1-12_AMi | REI1 | 161 | 167 | - | 13.40 | CCCCTGA |
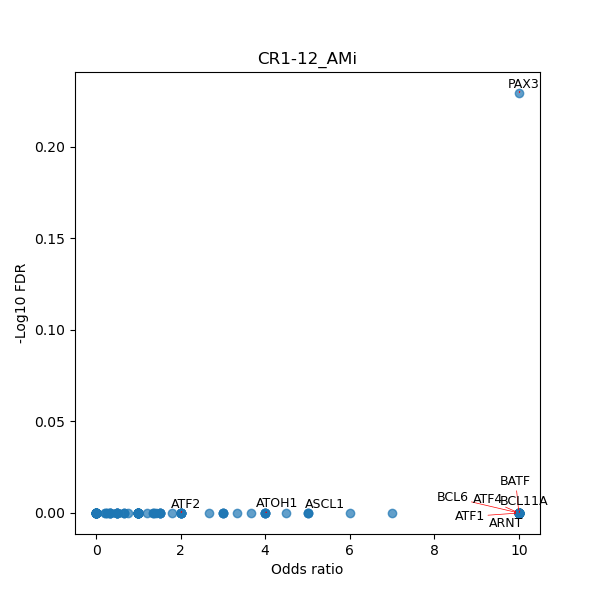
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.