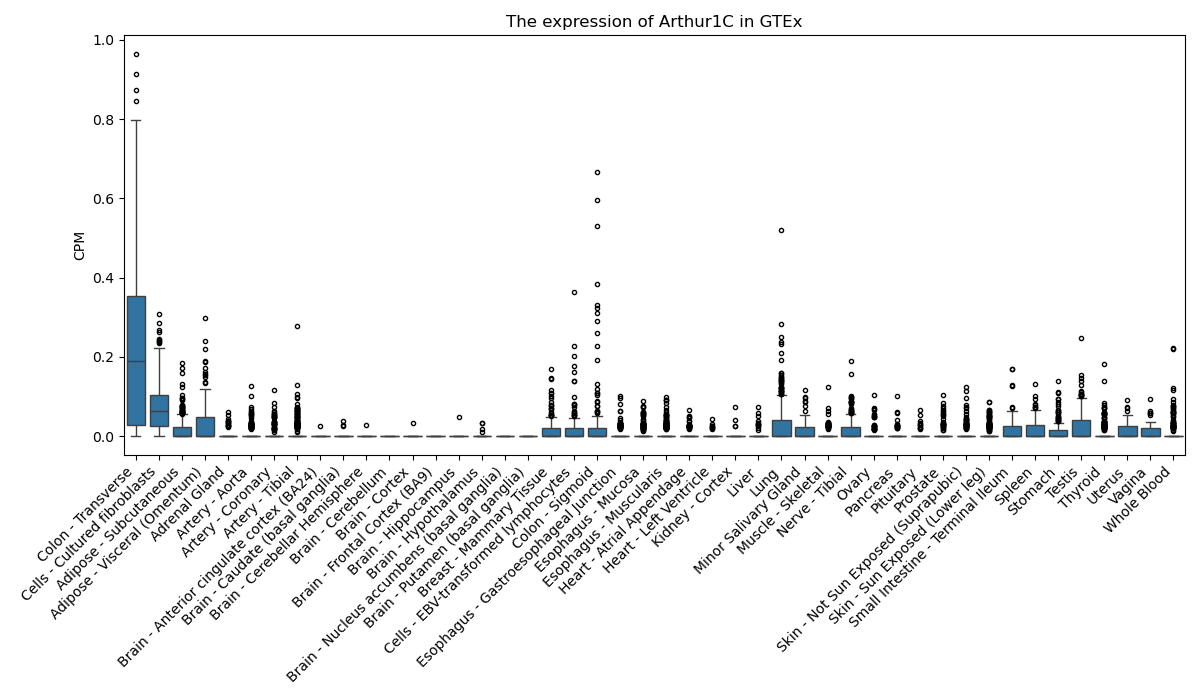
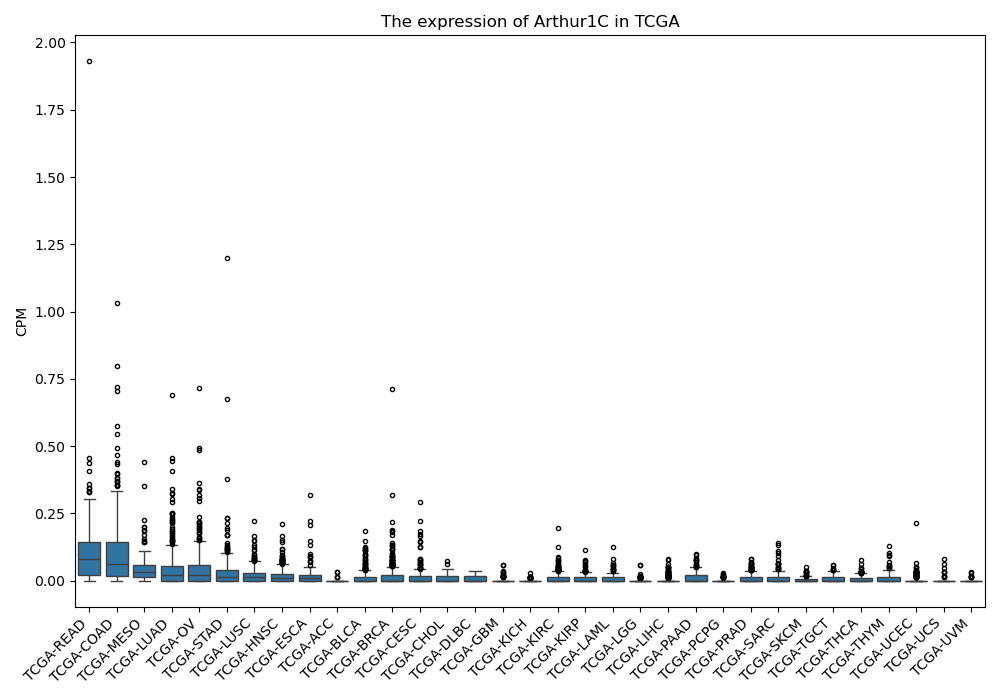
Arthur1C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000072 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 363 |
| Kimura value | 24.85 |
| Tau index | 0.9926 |
| Description | hAT-Tip100 DNA transposon, Arthur1C subfamily |
| Comment | Arthur1C has 8 bp TSDs. Positions 1-67 and 217-363 correspond to positions 1-67 and 3801-3947 of the autonomous Arthur1. Region 68-216 is unique to Arthur1C. |
| Sequence |
CAGAGGCGGATTTACCGTGAAGCTAATGAAGCTTAAGCTTCAGGGCCCCTCACTTGCACGGGCCCCTGTGAGTGCTGGGAGTTGCTGGGAGTTGTAGAGTGTTCTAGGTGGGGAGGGGAAGCCAGGTTGCAATCAGGAAGCATTTCTATGTAAGCATTTCTGGTAAATTGCCTAAAGAGATCTCAGAAGAAAGGGACCTGAATCTCCAAGGCTCCAGTAATTTGTTGTGATTTCTTTTCTCATTCTAAATAAATATTCACTTTCGTACCTAATTTTGTATTCGTAATTTTGTATTCTTTTTCTTAAAGAGGGCCCCCCAAATTGTATAAGCTTCAGGCCCCACAAAACCTGGATCCGCCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Arthur1C | Glyma19g26560.1 | 336 | 343 | + | 14.23 | GGCCCCAC |
| Arthur1C | GLYMA19G26560 | 336 | 343 | + | 14.23 | GGCCCCAC |
| Arthur1C | TB1 | 311 | 319 | + | 14.18 | GGCCCCCCA |
| Arthur1C | FOXD2 | 245 | 255 | + | 14.08 | CTAAATAAATA |
| Arthur1C | LjSGA_053525.1 | 336 | 343 | + | 14.06 | GGCCCCAC |
| Arthur1C | Rfx6 | 81 | 89 | - | 13.89 | CCCAGCAAC |
| Arthur1C | KLF14 | 110 | 118 | + | 13.88 | GGGGAGGGG |
| Arthur1C | FOXO1::FLI1 | 129 | 141 | + | 13.83 | GCAATCAGGAAGC |
| Arthur1C | GFI1 | 223 | 233 | - | 13.66 | AAATCACAACA |
| Arthur1C | PLAGL2 | 43 | 50 | + | 13.61 | GGGCCCCT |
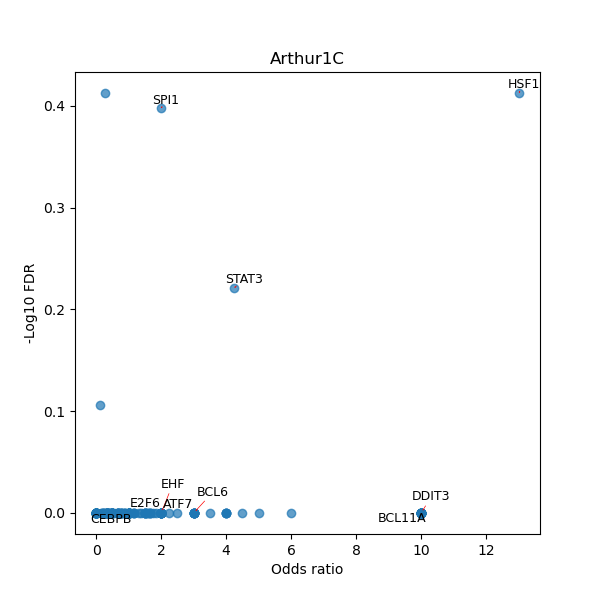
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.