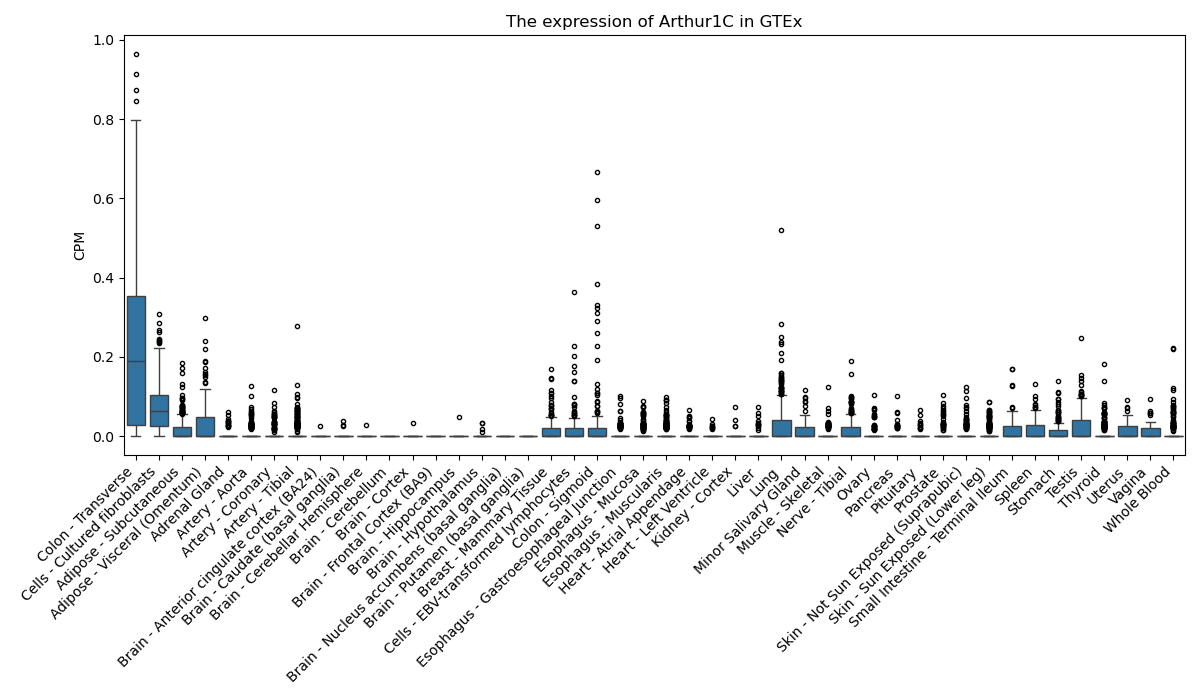
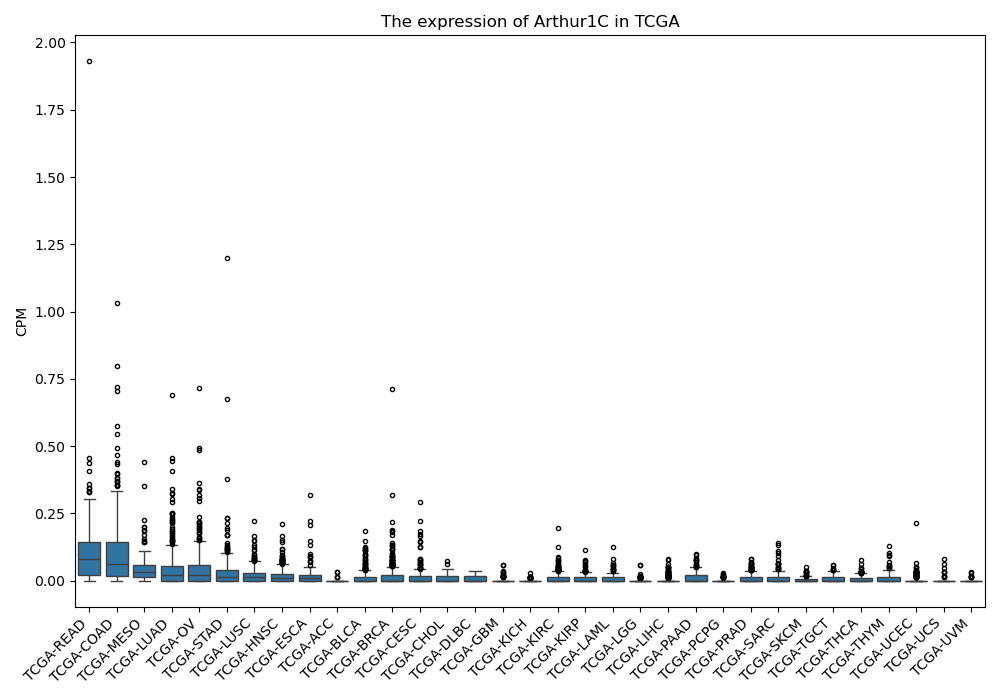
Arthur1C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000072 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 363 |
| Kimura value | 24.85 |
| Tau index | 0.9926 |
| Description | hAT-Tip100 DNA transposon, Arthur1C subfamily |
| Comment | Arthur1C has 8 bp TSDs. Positions 1-67 and 217-363 correspond to positions 1-67 and 3801-3947 of the autonomous Arthur1. Region 68-216 is unique to Arthur1C. |
| Sequence |
CAGAGGCGGATTTACCGTGAAGCTAATGAAGCTTAAGCTTCAGGGCCCCTCACTTGCACGGGCCCCTGTGAGTGCTGGGAGTTGCTGGGAGTTGTAGAGTGTTCTAGGTGGGGAGGGGAAGCCAGGTTGCAATCAGGAAGCATTTCTATGTAAGCATTTCTGGTAAATTGCCTAAAGAGATCTCAGAAGAAAGGGACCTGAATCTCCAAGGCTCCAGTAATTTGTTGTGATTTCTTTTCTCATTCTAAATAAATATTCACTTTCGTACCTAATTTTGTATTCGTAATTTTGTATTCTTTTTCTTAAAGAGGGCCCCCCAAATTGTATAAGCTTCAGGCCCCACAAAACCTGGATCCGCCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Arthur1C | TCP14 | 308 | 325 | - | -1.55 | ACAATTTGGGGGGCCCTC |
| Arthur1C | IRF9 | 253 | 267 | - | -4.13 | TACGAAAGTGAATAT |
| Arthur1C | ZAP1 | 14 | 28 | + | -4.55 | ACCGTGAAGCTAATG |
| Arthur1C | EWSR1-FLI1 | 113 | 130 | + | -10.29 | GAGGGGAAGCCAGGTTGC |
| Arthur1C | EWSR1-FLI1 | 109 | 126 | + | -15.82 | TGGGGAGGGGAAGCCAGG |
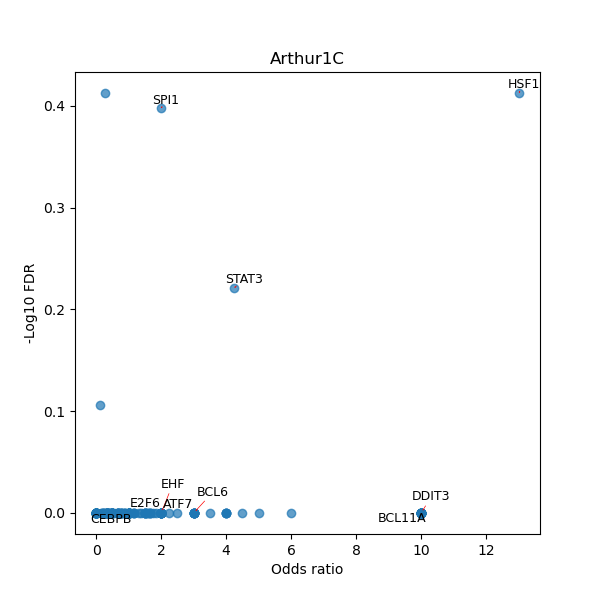
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.