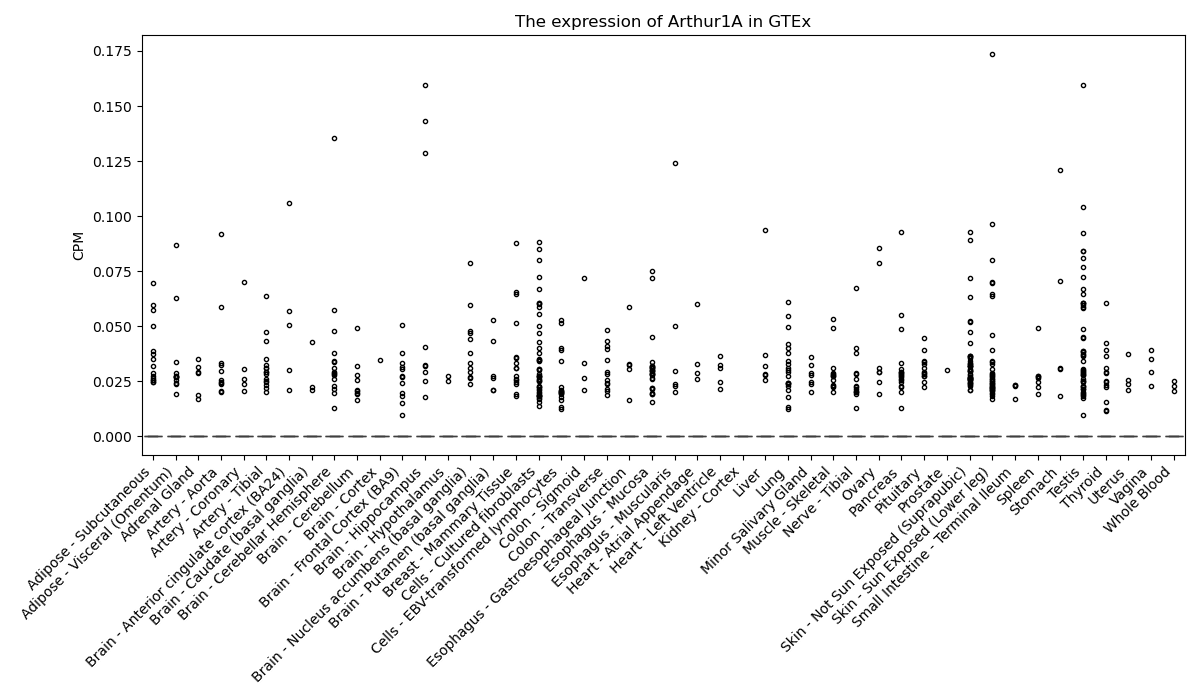
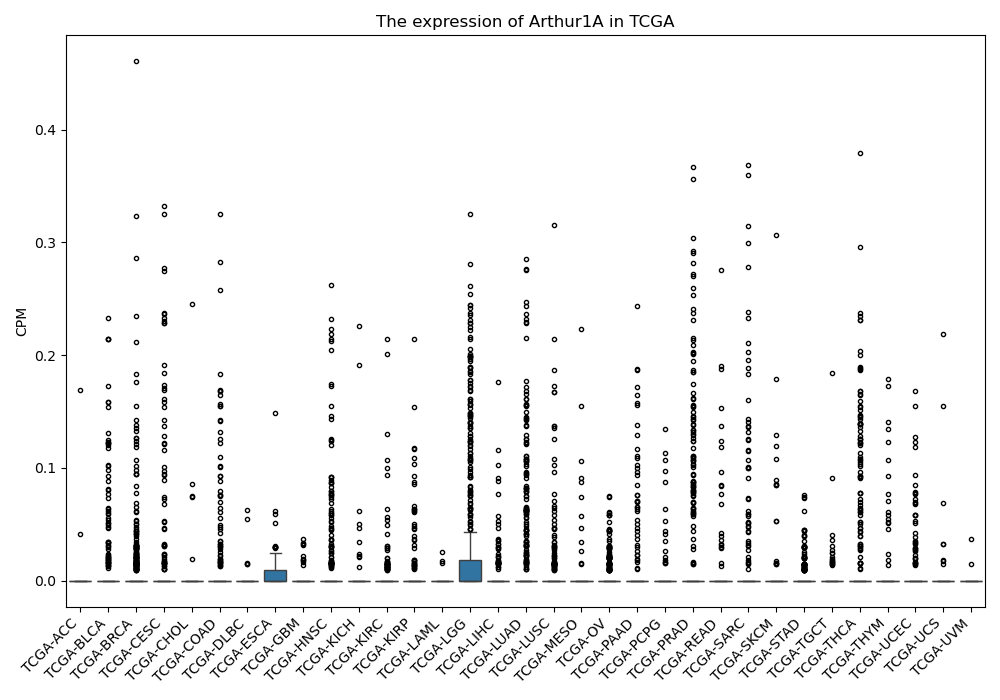
Arthur1A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000070 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 177 |
| Kimura value | 25.80 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Arthur1A subfamily |
| Comment | Arthur1A has 8 bp TSDs and 12 bp TIRs. Positions 1-79 and 79-177 correspond to 1-79 & 3849-3947 of the autonomous Arthur1. |
| Sequence |
CAGAGGCGGATTTACCGTGAAGCTAATGAAGCTTAAGCTTCAGGGCCCCTCACTTGCACGGGCCCCTTCCAAGGCCCTGTACCTAATTTTGTATTCGTAATTTTGTATTCTTTTTCTTAAAGAGGGCCCCCCAAATTGTATAAGCTTCAGGCCCCACAAAACCTGGATCCGCCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Arthur1A | MYB27 | 80 | 91 | - | 17.77 | CAAAATTAGGTA |
| Arthur1A | TB1 | 124 | 132 | + | 17.38 | GGGCCCCCC |
| Arthur1A | Prdm15 | 157 | 167 | + | 16.85 | CAAAACCTGGA |
| Arthur1A | PLAGL2 | 124 | 131 | + | 15.67 | GGGCCCCC |
| Arthur1A | MYB57 | 80 | 89 | + | 15.44 | TACCTAATTT |
| Arthur1A | ARALYDRAFT_495258 | 150 | 157 | + | 15.43 | GGCCCCAC |
| Arthur1A | ARALYDRAFT_484486 | 150 | 157 | + | 15.43 | GGCCCCAC |
| Arthur1A | PK19717.1 | 124 | 132 | + | 15.24 | GGGCCCCCC |
| Arthur1A | MYB108 | 80 | 89 | + | 14.75 | TACCTAATTT |
| Arthur1A | ARALYDRAFT_493022 | 150 | 157 | + | 14.64 | GGCCCCAC |
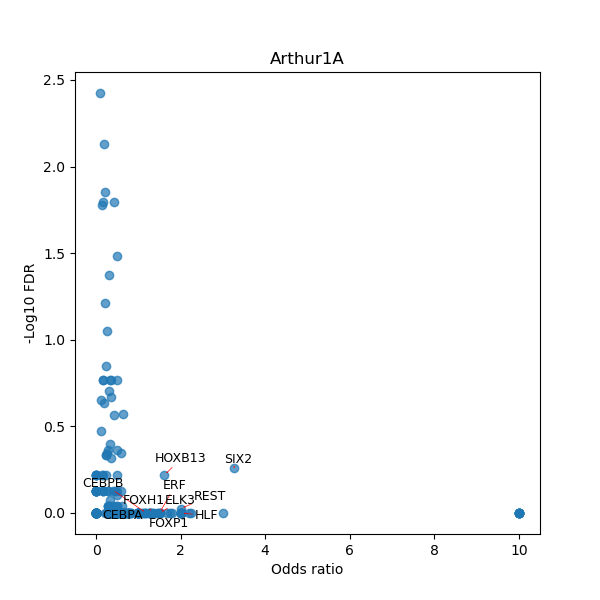
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.